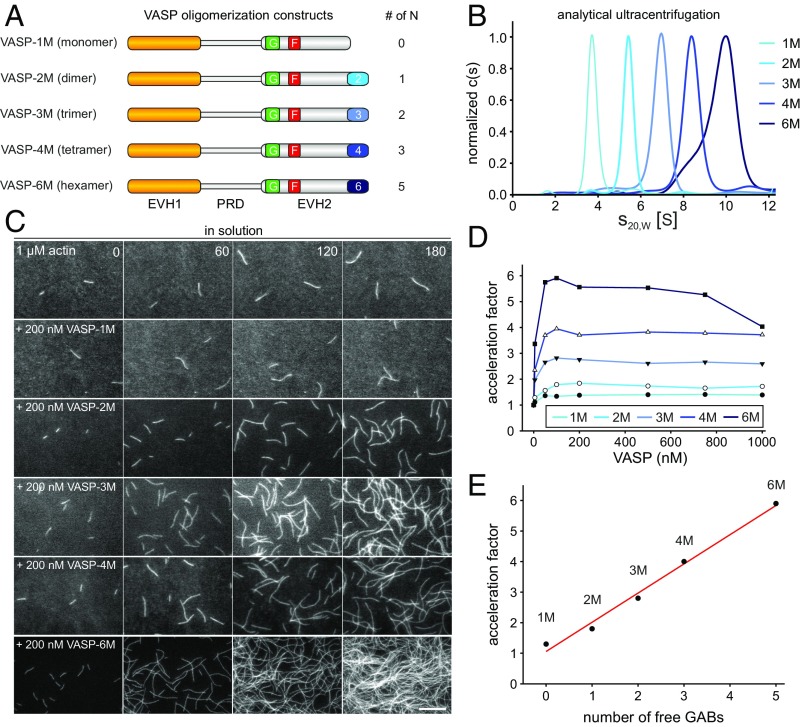
Fig. 1.
VASP-mediated filament elongation correlates directly with N in solution. (A) Schematic domain organization of the generated VASP-oligomerization mutants. Abbreviations: EVH1, Ena/VASP-homology domain 1; PRD, proline-rich domain; EVH2, Ena/VASP-homology domain 2 containing the G-actin-binding site (GAB) from Dictyostelium, the F-actin binding site (FAB), and the oligomerization domain. Abbreviations: F, FAB; G, GAB. (B) Sedimentation velocity analysis of the used VASP-oligomerization mutants fused to MBP. Differential sedimentation coefficient distribution analysis showed the proteins to sediment with s20°,w = 4.0 S (VASP-1M), 5.7 S (VASP-2M), 7.3 S (VASP-3M), 8.8 S (VASP-4M), and 9.9 S (VASP-6M) and is consistent with their predicted oligomerization status. (C) Time-lapse micrographs of TIRFM assays—corresponding to Movie S1—for determination of elongation rates. The actin (1 μM, 23% Alexa 488 labeled) was polymerized in the absence or presence of 200 nM of the indicated VASP construct in TIRF buffer, respectively. Time is given in seconds. (Scale bar, 20 µm.) (D) Dependence of the acceleration of actin-filament elongation on the oligomerization state of VASP. The elongation rates derived from TIRFM movies as shown in C were divided by the rate of spontaneous actin assembly to obtain the acceleration factors for each VASP oligomerization construct at the concentrations indicated. Concentrations are given in monomers. Fifteen actin control filaments and 30 filaments for all other data points in presence of VASP were measured. (E) Acceleration of VASP-mediated actin assembly at 100 nM VASP oligomer increased as a function of N (=total GABs − 1) as predicted by the kinetic model (9). Red line shows linear fit.