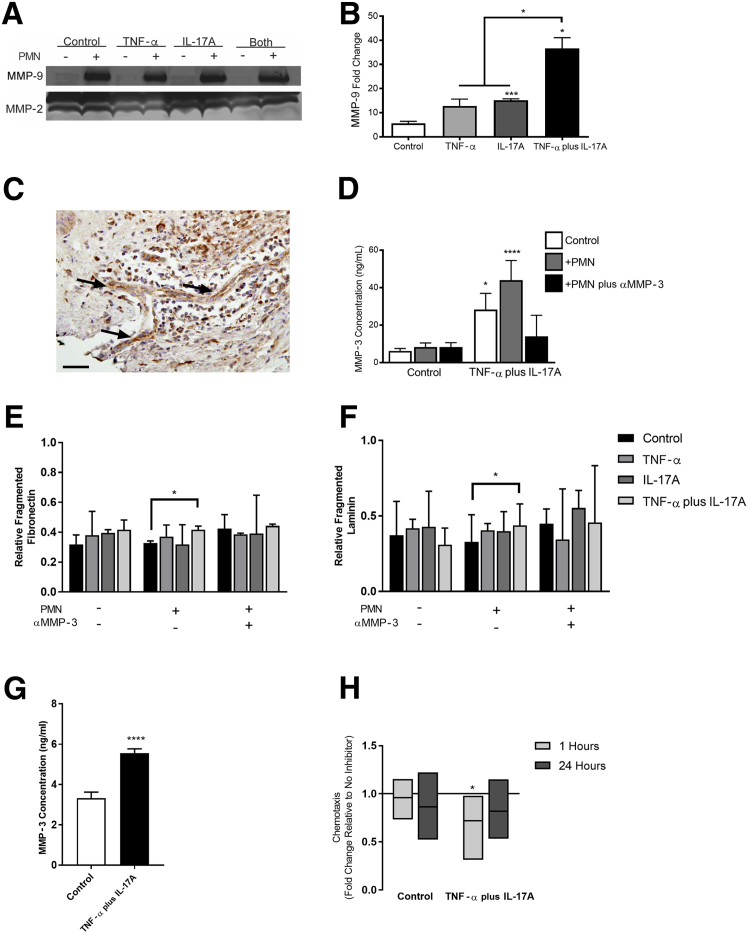
Figure 6.
Neutrophils and pericytes (PCs) mediate basement membrane proteolysis. A: Gelatin zymography results for active matrix metalloproteinase (MMP)-9 and MMP-2 in PC culture media after 5 days of activation and 24 hours of incubation with (+) or without (−) neutrophils. B: Active MMP-9 in cell culture media with neutrophils (24 hours); data are represented as fold change over the baseline level found in each PC protein condition without neutrophils. C: Representative histologic image of MMP-3 staining in human Sweet syndrome; Arrow indicates postcapillary venule. D: MMP-3 expression in PC cell culture media after 5 days of activation and 24 hours of incubation with or without neutrophils, as determined by enzyme-linked immunosorbent assay (ELISA) [one-way analysis of variance relative to control with no polymorphonuclear leukocytes (PMN)]. The ratio of fragmented to intact fibrillar fibronectin (E) and fibrillar laminin (F) in PC basement membrane after 5 days of activation and 24 hours of incubation with or without neutrophils. Data were standardized to total protein with a bicinchoninic acid assay. Bars represent median with interquartile range. G: MMP-3 expression of neutrophils in contact with decellularized PC media for 1 hour, as determined by ELISA. H: Neutrophil chemotaxis along an IL-8 gradient through decellularized PC protein after 5 days of activation in culture; neutrophils were treated with an anti–MMP-3 inhibitor, and chemotaxis is shown relative to controls without inhibitor. Bars represent maximum, mean, and minimum. H: MMP-3 expression of neutrophils in contact with decellularized PC media for 1 hour, as determined by ELISA. The solid line indicates no change compared with the same condition without an MMP-3 inhibitor. Data are expressed as means ± SEM (B, D, and G). Bars represent medians with interquartile ranges (E and F). Bars represent means ± minimum/maximum (H). n = 3 (B); n ≥ 2 (D, each run in quadruplicate); n ≥ 3 (E and F); n = 3 (G, each run in quadruplicate); n ≥ 3 (H). ∗P < 0.05, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001. Scale bar = 100 μm (C).