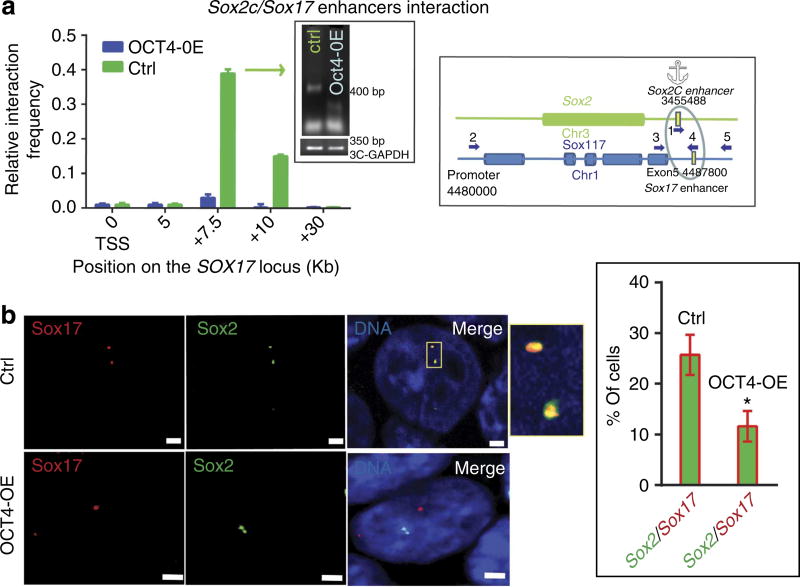
Figure 7. OCT4 breaks Sox-2/Sox-17 enhancer inter-chromosomal interaction.
(a) 3C experiments were performed on digested and religated chromatin from undifferentiated ZHBTc6 cells or OCT4-induced mesendodermal cells. Primers spanning within the Sox-2 and Sox-17 loci and numbered as in Table 2 were used in PCR. Control primers, such as in exons, were used as controls (cartoons, a–c). Interaction frequencies were normalized using the GAPDH locus. The graphs show the frequency of interactions between chromosomes (Chr) 1 and 3. The gels show the specificity of the amplicons of the peaks of interaction; the sequence of amplicons was checked by sequencing. The relative interaction frequency value is set to one for the interaction measured between the control GAPDH fragments. Data are representative of four experiments (mean ± s.e.m.). (b) Illustration of FISH experiments using a Sox2alexa488 and Sox17alexa568 probes showing the interaction between chromosomes 1 and 3 for both alleles (the nucleus is representative of 116 imaged nuclei of pluripotent stem cells; upper panels). Alleles of chromosomes 1 and 3 were separated in mesendodermal OCT4-OE cells (the nucleus is representative of 110 nuclei; lower panels). The red, green channels and the merged image are shown. Scale bar, 2 µm. The probe hybridized region is magnified in the inset of the merged image. The right graph in the inset indicates the averaged percentage of cells featuring a Sox-2/Sox-17 interaction from three experiments (*Student’s t-test, P≤0.01)