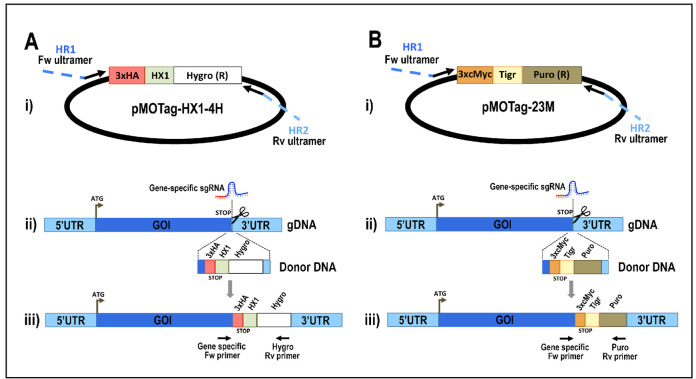
Figure 5. Schematic representation of strategies used to generate endogenous C-terminal tagging in Trypanosoma cruzi .
. A. i) pMOTag-HX1-4H vector map. The T. cruzi HX1 trans-splicing signal is located between the 3xHA tag sequence and the gene that confers resistance to hygromycin (Hygro R). HR1 Fw and HR2 Rv ultramers indicate oligonucleotides used to amplify DNA donor cassette. The annealing regions for ultramers to pMOTag-HX1-4H and genomic DNA (gDNA) are indicated in black and blue, respectively. ii) A double-stranded gDNA break was produced by Cas9 targeted by the sgRNA both expressed from 3’end-sgRNA/Cas9/pTREX plasmid downstream of the STOP codon of the gene of interest (GOI) in the endogenous locus. Homologous directed repair was induced co-transfecting epimastigotes with the DNA donor cassette, containing homologous regions to the GOI 3’ end (blue) and to the GOI 3’UTR (light blue). iii) Integration of 3xHA and antibiotic resistance gene at 3’end of GOI by homologous recombination. Arrows indicate primers used for checking integration of donor DNA. B. i) pMOTag23M vector map. The 3xc-Myc tag sequence and the puromycin resistance gene (Puro R) are separated by the T. brucei tubulin intergenic region (Tigr). The rest of i), ii) and iii) description is similar to that panel A. Hygro, hygromycin resistance gene; Puro, puromycin resistance gene; UTR, 5’ and 3’ untranslated regions; ATG, start codon. This figure was originally published in Lander et al., 2016b .