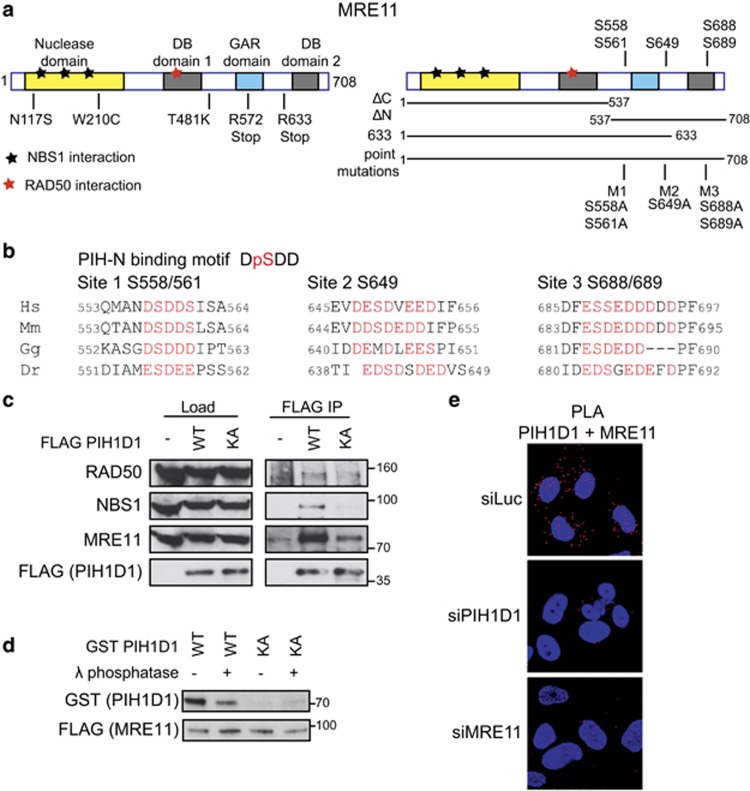
Figure 1.
PIH1D1 interacts with MRN complex. (a) Schematic representation of human MRE11 comprising of a nuclease domain, two DNA-binding domains (DB1 and DB2) and glycine–arginine-rich (GAR) domain. Positions of ATLD mutations (left panel) and putative PIH-N phospho-binding sites (right panel) are indicated. Bars below the right panel represent the MRE11 constructs used in this study. (b) Conservation of the PIH-N consensus motif in the C terminus of MRE11. Acidic amino acids and serines that form potential PIH-N-binding sites are displayed in red. (c) FLAG PIH1D1 WT interacts with MRN complex components. HEK293T cells transfected with FLAG PIH1D1 WT or phospho-binding mutant PIH1D1 KA were lysed in immunoprecipitation (IP) buffer (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 2.5 mM EGTA, 10% (v/v) glycerol supplemented with cOmplete EDTA-free protease inhibitor, PhosSTOP phosphatase inhibitor (Sigma-Aldrich, St Louis, MO, USA) and EtBr (50 μg/ml)) and sonicated 3 × 10s. Cleared cell extracts were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 h. Beads were washed 4x with IP buffer, boiled in 2xLSB buffer (100 mM Tris (pH 6.8), 200 mM dithiothreitol (DTT), 4% sodium dodecyl sulfate (SDS), 0,2% bromophenol blue, 20% (v/v) glycerol) and bound proteins were detected by immunoblotting using antibodies against RAD50 (Abcam, Cambridge, UK; ab119708), MRE11 (Cell Signaling, Danvers, MA, USA; no. 4895) and NBS1 (Cell Signaling; no. 3002). (d) MRE11–PIH1D1 interaction depends on MRE11 phosphorylation and on the PIH-N domain of PIH1D1. FLAG MRE11 was purified from HEK293T cells using anti-FLAG M2 affinity gel. Beads were washed with IP buffer supplemented with 1 m NaCl, treated or not with λ phosphatase for 30 min at 30 °C and MRE11 was eluted with 3 × FLAG peptide (30 μg; Sigma-Aldrich). GST PIH1D1 WT and phospho-binding mutant GST PIH1D1 KA were purified from BL21 Escherichia coli and pull-down with purified MRE11 was performed as described previously.20 (e) MRE11 and PIH1D1 interact in vivo. U2OS cells grown on coverslips were transfected with 40 nm siRNA targeting luciferase (5′-CGUACGCGGAAUACUUCGA-3′ Sigma), PIH1D1 (1:1 mixture of 5′-GAAUGGAAAUGUAGUCUUA-3′ and 5′-GAGAAGAGGCUGCUGGCUU-3′ in the untranslated region of PIH1D1 as described20) or MRE11 (5′-GAGCAUAACUCCAUAAGUA-3′ in the untranslated region of MRE11 mRNA) with Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's instructions. Cells were permeabilized with 0.2% Triton X-100 for 5 min at room temperature and proximity ligation assay (PLA) was performed using MRE11 (Cell Signaling; no. 4895), PIH1D1 (Abcam; ab57512) antibodies and Duolink reagent (Sigma-Aldrich) according to the manufacturer's protocol. Red signal is proximity ligations assay (PLA) staining, and 4',6-diamidino-2-phenylindole (DAPI) is in blue. Representative image is shown.