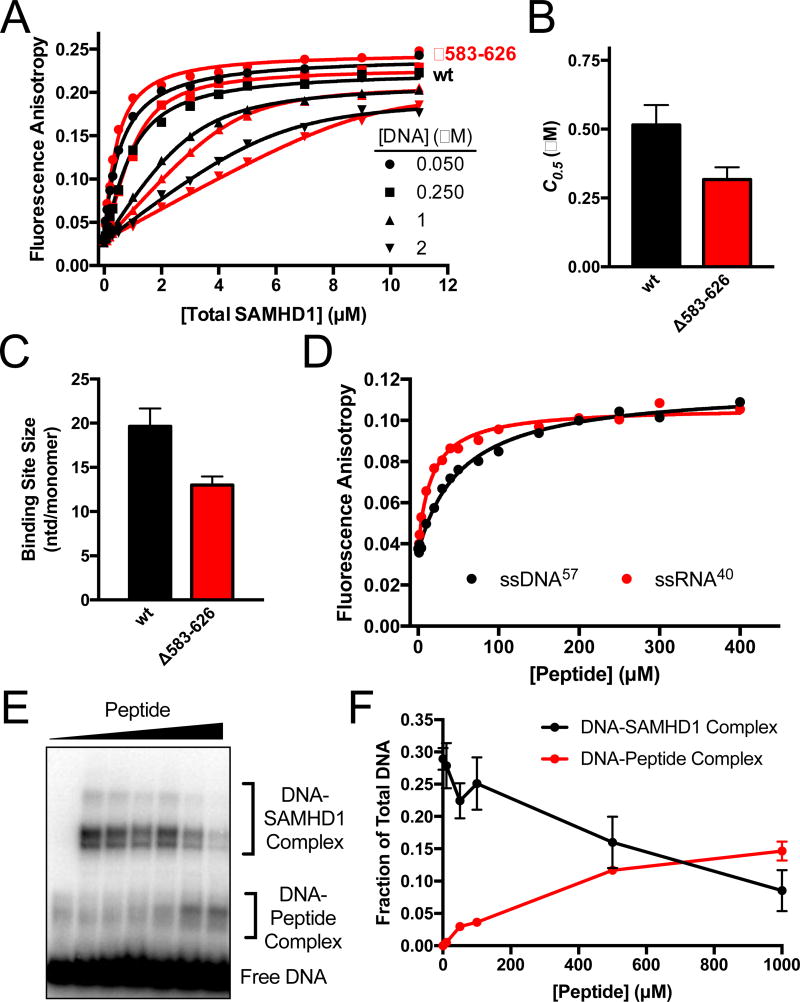
Figure 5. The disordered C-Terminus of SAMHD1 binds ssNA.
(A) Fluorescence anisotropy titrations of the fluorescein-labeled dT60 with wt and Δ583–626 SAMHD1 as a function of DNA concentration. The data were fit to a variable stoichiometry quadratic binding equation17. (B) Comparisons of the concentrations of wt and Δ583–626 SAMHD1 required for half-maximal DNA binding (C0.5). (C) Comparison of the calculated binding site sizes of wt SAMHD1 and Δ583–626 SAMHD1. The sizes are indicated as nt/monomer. (D) Fluorescence anisotropy increases of FAM-dT60 and FAM-ssRNA40 upon binding of a peptide consisting of residues 582–626 of SAMHD. (E) BrdU crosslinking reactions were performed with fixed concentrations of BrdU DNA (5 µM) and SAMHD1 (5 µM), and variable concentrations (0 to 1 mM) of the 582–626 SAMHD1 peptide. (F) Quantification of the DNA-SAMHD1 and DNA-peptide complexes from (F) by densitometry.