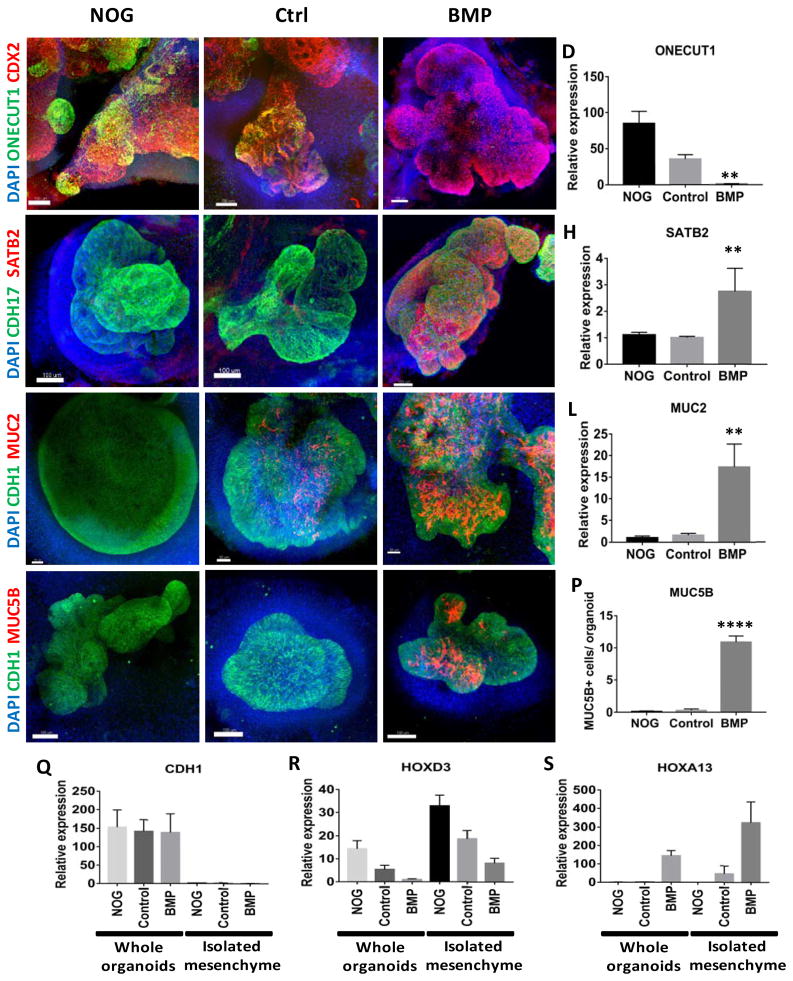
Figure 3.
Regional patterning is maintained in human intestinal organoids following prolonged in vitro culture.
(A–D) Whole-mount immunofluorescence and QPCR analysis with the proximal marker ONECUT1 (green) of 28 day old organoids that resulted from the initial 3 day treatment of spheroids with NOGGIN, control, or BMP2. Staining with CDX2 (red) and DAPI (blue) were also used to detect the epithelium and mesenchyme. (E–H) Expression of the posterior marker SATB2 (red) detected by IF and by QPCR. (I–L) Analysis of the pan-goblet cell marker MUC2 (red) by IF and by QPCR. (M–P) Analysis of the colon-specific goblet cell marker MUC5B (red) by IF. The number of MUC5B+ cells was quantified in (P). (Q–S) Analysis of patterning markers in isolated mesenchyme cultures relative to whole organoids. QPCR analysis of CDH1 (Q), the proximal HOX gene HOXD3 (R), and the distal HOX gene HOXA13 (S) in whole organoids and in mesenchyme cultures derived from NOGGIN, control, or BMP2 treated organoids. CDH1 was only observed in whole organoids that contained epithelial cells. Error bars represent SEM. For IF minimum of 10 organoids from at least 3 different experiments were examined for each condition. For QPCR a minimum of 5 biological replicates from 2 separate experiments were examined. Scale bars = 100 μm. **p ≤ 0.01 and ****p ≤ 0.0001 determined by 2 tailed t-test comparing NOGGIN+Control treated organoids and BMP2 treated organoids.