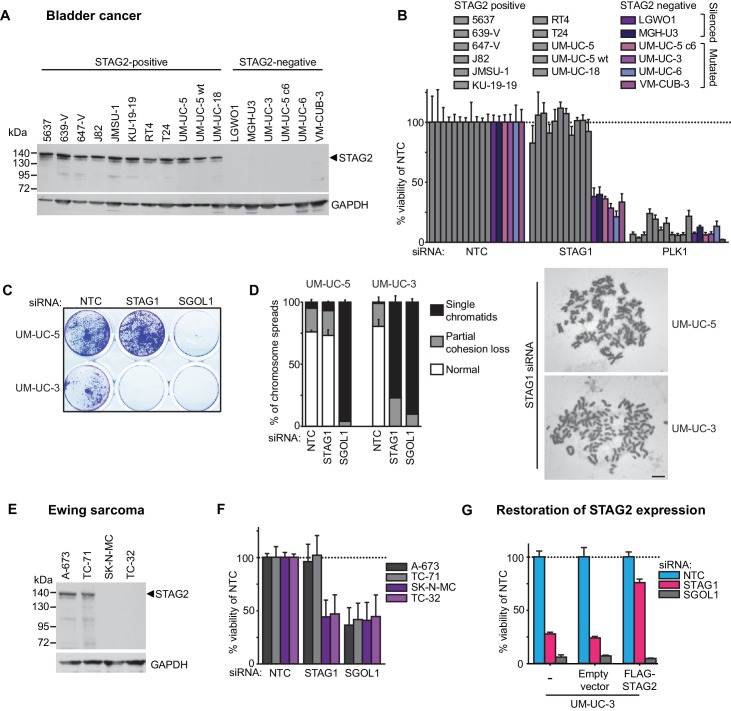
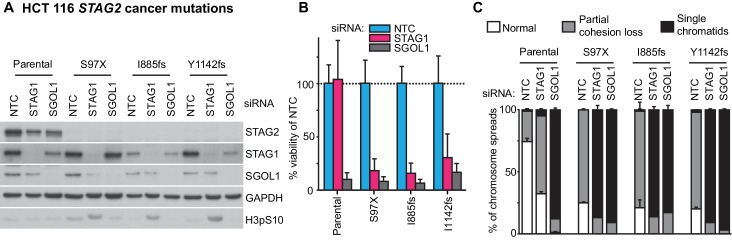
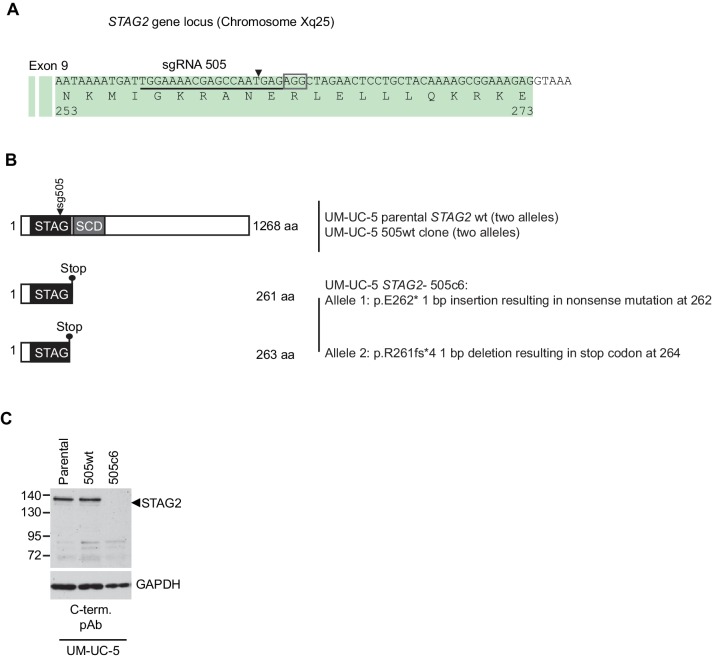
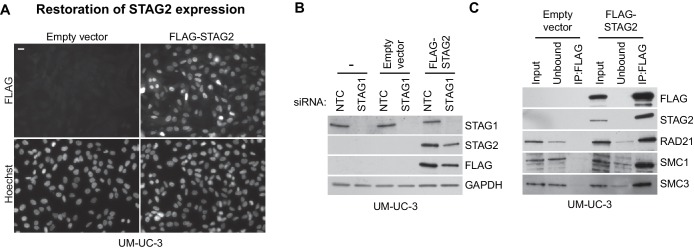
Figure 3. The synthetic lethal interaction between STAG1 and STAG2 is manifested in disease-relevant bladder cancer and Ewing sarcoma cell lines.
(A) The indicated bladder cancer cell lines were analyzed for STAG2 expression by immunoblotting. (B) The indicated bladder cancer cell lines were transfected with NTC, STAG1 and PLK1 siRNA duplexes. Viability was determined 7 or 10 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 2 independent experiments with 5 biological repeats each, error bars denote standard deviation). (C) STAG2 wild-type UM-UC-5 and STAG2 mutated UM-UC-3 cells were transfected with NTC, STAG1 and SGOL1 siRNA duplexes. Colony formation was analyzed 7 days after transfection by crystal violet staining. (D) 72 hr after siRNA transfection into UM-UC-5 and UM-UC-3 cells, Giemsa-stained chromosome spreads were prepared and analyzed for sister chromatid cohesion phenotypes (n = 100 spreads, error bars denote standard deviation of two independently analyzed slides). (E) The indicated Ewing sarcoma cell lines were analyzed for STAG2 protein expression by immunoblotting. (F) The indicated Ewing sarcoma cell lines were transfected with NTC, STAG1 and SGOL1 siRNA duplexes. Viability was measured 6 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 3 independent experiments with 3 biological replicates each, error bars denote standard deviation). (G) STAG2 mutated UM-UC-3 cells were transduced with a lentivirus encoding a FLAG-STAG2 transgene. Stably selected cell pools were subsequently transfected with NTC, STAG1 or SGOL1 siRNA duplexes. Viability was measured 7 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 4 biological replicates, error bars denote standard deviation).