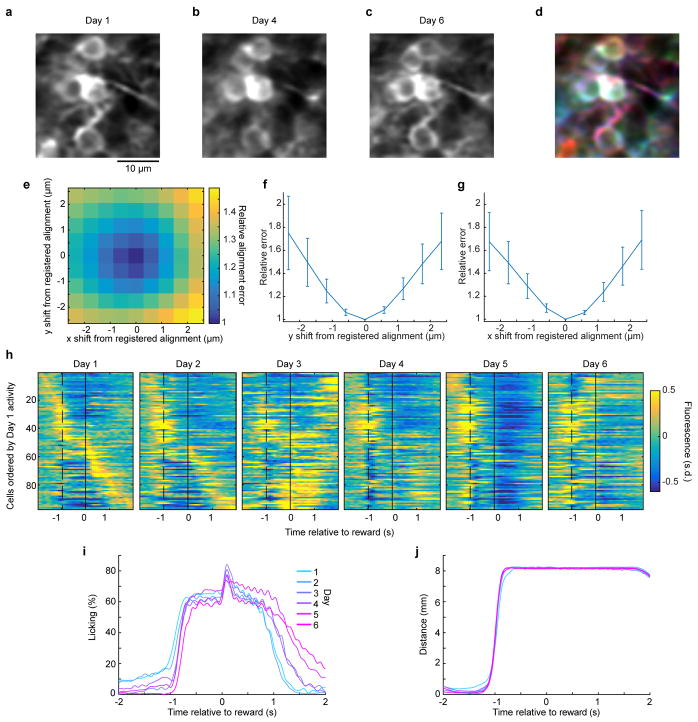
Extended Data Fig. 9. Chronic imaging cell tracking and registration.
a–c, Magnified view of mean two-photon image from the regions shown in Fig. 4a on Day 1 (a), Day 4 (b), and Day 6 (c). d, Colorized overlay of the images in a–c in red, blue and green. We rigidly aligned the mean fluorescence image on each day to that of the final day using TurboReg37, resulting in unambiguous alignment of visible morphological features of individual granule cells. e, To quantify any ambiguity in the image registration we offset our images from optimal alignment by small amounts. For one example session, we quantified the image concordance of Day 1 and Day 6, as a function of displacing the Day 1 image in the x and y directions relative to the registered optimum at zero (sum squared pixel differences between days, normalized to the registered optimum). There is a clear trough in the alignment error at the optimum, demonstrating that even slight, submicron misalignments are easily detected by image registration. Thus, there is little appreciable ambiguity in the alignment procedure. f, g, Average alignment error as a function of image displacement from the registered optimum, as in e, here averaged across all sessions and mice (n = 15 alignments from 3 mice). Error bars denote s.e.m. across alignments. Even the smallest, submicron, single pixel displacements result in significantly higher alignment error than the registered optimum (p = 4.4 × 10−6 and 5.8 × 10−5 for one-pixel x and y misalignments respectively, Wilcoxon signed-rank test). h, Mean fluorescence response of all neurons for the example mouse shown in Fig. 4c, here ordered by their Day 1 activity peak response time (n = 97 neurons). i–j, Change over the 6 days of the imaging study in licking behavior (i) and forelimb movement behavior (j) for the mouse in (h). Gross changes in motor behavior were relatively modest over the days of the imaging study (Methods).