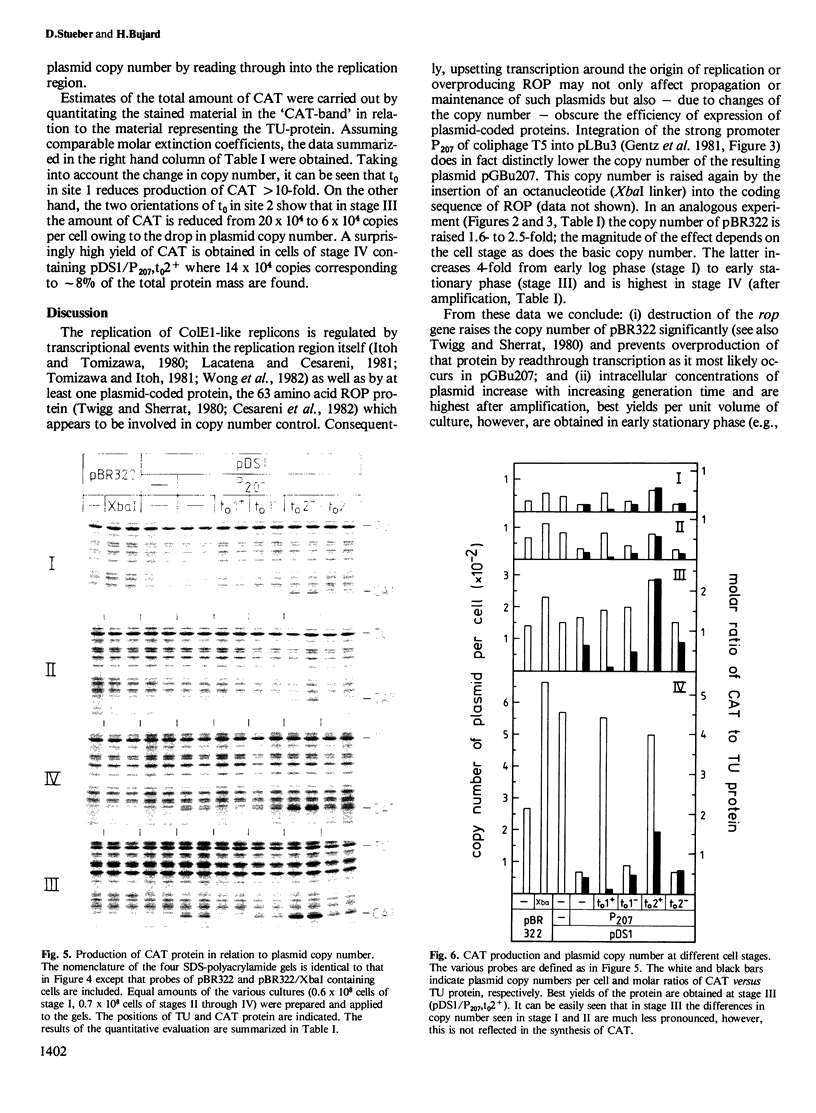
Abstract
The copy number of plasmids containing the ColE1 replicon is affected by changes in the transcriptional activity within the plasmid if these changes lead to transcriptional readthrough into the replication region towards the promoter priming DNA replication. Such readthrough e.g., from the tet region in pBR322 not only causes overproduction of a peptide known to affect the copy number negatively but also appears to interfere negatively with the replication of the plasmid itself. The proper placement of efficient transcriptional terminators prevents such interference and permits the stable integration of strong promoters. Due to this termination effect, up to 9-fold differences in plasmid copy number were observed, depending upon the particular growth conditions. The higher copy number is of course reflected by higher yields of plasmid-specified gene products indicating the relevance of the above effects for studies of gene expression.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K., Clark B. F., Duffy L., Jones M. D., Kaziro Y., Laursen R. A., L'Italien J., Miller D. L., Nagarkatti S., Nakamura S. Primary structure of elongation factor Tu from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Langner A., Chang A. C., Cohen S. N., Bujard H. Cloning and analysis of strong promoters is made possible by the downstream placement of a RNA termination signal. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4936–4940. doi: 10.1073/pnas.78.8.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981 Dec 17;294(5842):623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Bloch P. L., Reeh S., Neidhardt F. C. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978 May;14(1):179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Packman L. C., Burleigh B. D., Dell A., Morris H. R., Hartley B. S. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979 Dec 20;282(5741):870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Stüber D., Delius H., Bujard H. Electron microscopic analysis of in vitro transcriptional complexes: mapping of promoters of the coliphage T5 genome. Mol Gen Genet. 1978 Oct 30;166(2):141–149. doi: 10.1007/BF00285916. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Wong E. M., Muesing M. A., Polisky B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]