Fig. 1.
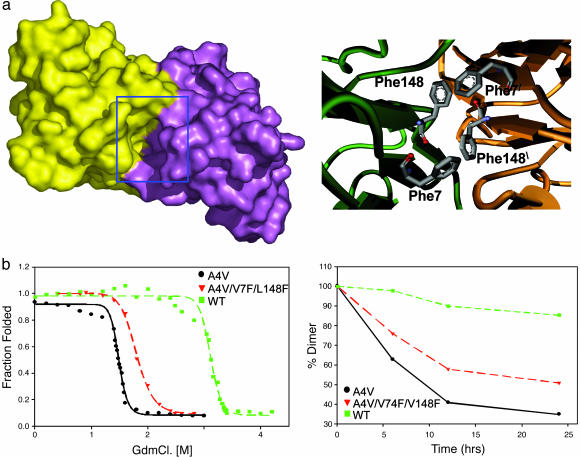
When the cavity at the SOD1 dimer interface is partially filled by mutagenesis, the resultant protein is more stable and aggregates more slowly. (a Left) shows a surface representation of A4V mutant SOD1 dimer colored to show the two subunits. A deep cavity at the dimer interface is highlighted by the blue box. The surface was generated by using a water molecule as a probe. (a Right) A model of the SOD1 cavity in the variant A4V/V7F/V148F. Note the quartet of phenylalanine sidechains at the dimer interface. (b Left) GdnCl unfolding of A4V (black), WT (green) and the A4V/V7F/V148C triple mutant (red) plotted as fraction unfolded vs. GdnCl concentration. (b Right) Loss of SOD1 dimers over time parallels the formation of aggregates (data not shown). The rate of aggregation is inversely correlated to the stability toward denaturation: A4V (black), WT (green), and A4V/V7F/L148C (red).