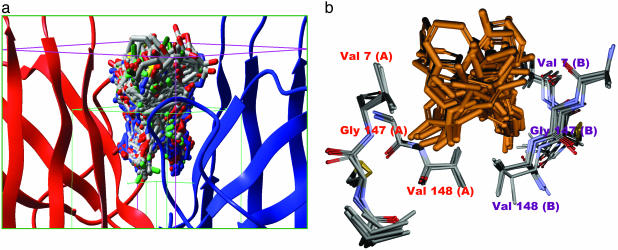
Fig. 2.
The protein and small-molecule portions of the docked complexes occupy overlapping space. (a) The grid box used for our docking calculation. The green box represents the conformational search space for the ligand; the pink box represents the boundary conditions used for the docking calculation. One hundred of the 2,000 final poses obtained from docking are shown docked at the dimer interface. (b) Superposition of five SOD1 x-ray structures WT, apo-WT, S134N, H46R, and A4V (Protein Data Bank codes: 1SPD, 1HL4, 1N19, 1OEZ, and 1UXL), showing critical residues of the dimer interface cavity used in docking calculations. The mean rms deviation (Cα)is <0.6 Å, indicating that the dimer interface is rigid in nature, which suggests that docked molecules should bind to all mutants. A superposition of 20 of the top 100 docked molecules is also shown.