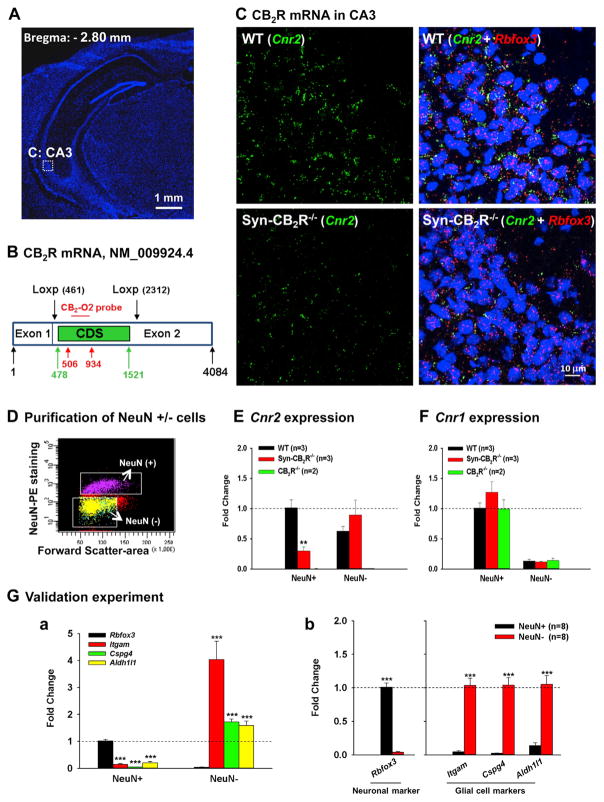
Figure 4. Neuronal CB2R mRNA Expression in the Hippocampus by RNAscope ISH and FACS-qPCR Assays.
(A) Hippocampal image (DAPI staining) illustrates the target region (CA3) in (C).
(B) The CB2R mRNA structure in CB2R-floxed mice and the target gene region of a CB2R RNAscope probe (CB2-O2, 506–934 bp) used to detect CB2R mRNA. The CB2-O2 probe targets the floxed region of mouse CB2R mRNA (NM_009924.4) in CB2R-floxed mice. CDS, (CB2R)-coding DNA sequence (478–1,521 bp).
(C) CB2R mRNA staining illustrates significant CB2R (Cnr2, green) and NeuN (Rbfox, red) mRNA co-localization in WT hippocampal CA3 neurons (upper panels), while such co-localization is substantially diminished in Syn-CB2R−/− (lower panels).
(D) A representative image shows FACS-sorted NeuN+ neurons and NeuN− non-neuronal cells in the hippocampus.
(E) The qPCR assays show that CB2R mRNA is detected mainly in NeuN+ hippocampal cells of WT mice, while the CB2R mRNA in NeuN+ hippocampal cells in Syn-CB2R−/− mice was substantially reduced (~70% reduction), and abolished in the CB2R−/− mice.
(F) The qPCR assays for CB1R mRNA (as controls) in the same samples demonstrate similar levels of CB1R mRNA expression in NeuN+ neurons and NeuN− cells in WT, Syn-CB2R−/−, and const. CB2R−/− mice.
(G) The qPCR assay results of neuronal and glial markers in two cell populations to examine the purity of sorted cells, illustrating that Rbfox3 was detected mainly in FACS-sorted NeuN+ neurons (a), but not in NeuN− cells (b). In contrast, the glial marker genes Itgam, Cspg4, and Aldh1l1 were mainly detected in NeuN− nonneuronal cells (b), but not in NeuN+ hippocampal neurons (a). Data shown in (a) were normalized to Rbfox3 expression in the NeuN+ population, which was defined as 1. Data shown in (b) were normalized to each respective marker gene level in NeuN+ (Rbfox3) and NeuN− cells (all three glial markers).