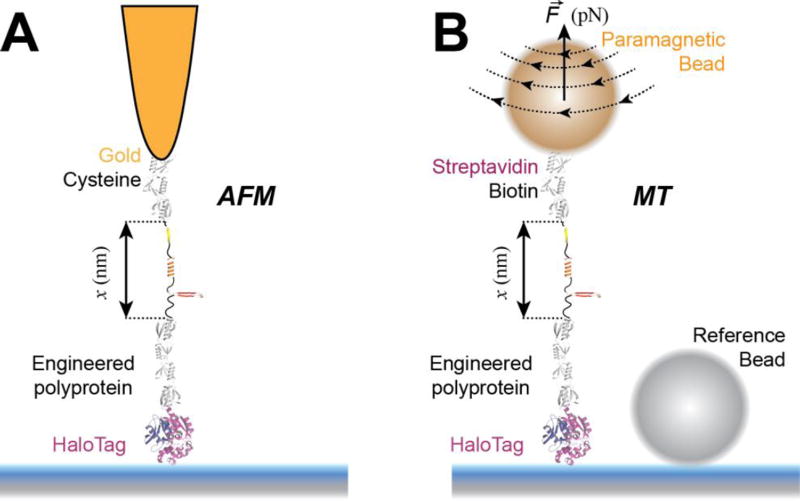
Figure 1. Single molecule force spectroscopy techniques used to study protein folding.
A) Schematics of an atomic force microscopy (AFM) experiment. A multidomain protein construct is covalently attached to the surface using HaloTag chemistry. A gold-coated cantilever with a tip having a radius of ~10 nm is used to pull the construct from the opposite end using gold-thiol attachment. Denaturation of protein domains leads to unfolding steps in the measured extension (in force-clamp mode) or to peaks in the measured force (in force-extension mode). B) Schematics of a magnetic tweezers (MT) experiment. A multidomain protein construct is covalently attached to the surface using HaloTag chemistry and tethered to a paramagnetic bead using the biotin-streptavidin bond. A reference non-magnetic bead glued to the glass surface is used to correct for drift. Denaturation of protein domains leads to unfolding steps in the measured extension. AFM is ideal for measuring fast occurring processes, such as unfolding of proteins at high forces, while MT excels on measuring slow-occurring events, such as unfolding and refolding of protein domains at low forces, taking place on a minute-to-hour time scale.