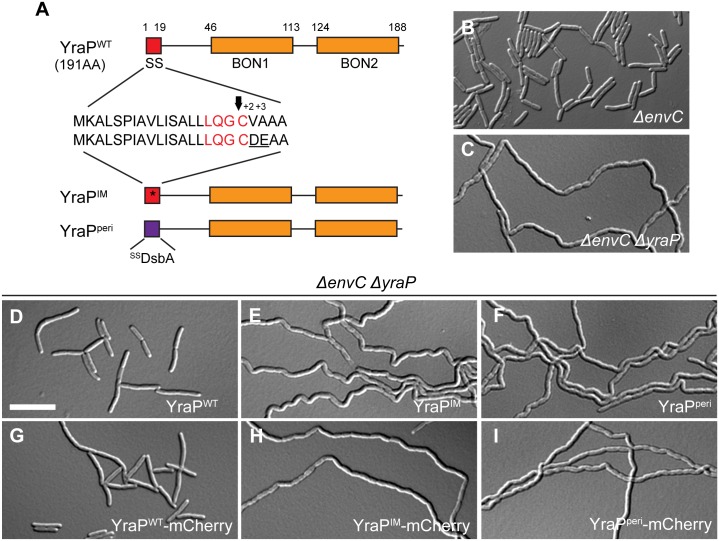
Fig 7. OM localization of YraP is required for cell separation in the absence of EnvC.
(A) The domain structure of YraP is illustrated. Indicated are the signal sequence (SS; red) and the two bacterial OsmY and nodulation domains (BON1/2; orange). Details of the signal sequence are presented with the lipobox in red and the arrow indicating the cleavage site just before the acylated cysteine. The IM-retained variant (YraPIM) contains a mutated signal sequence (indicated by the asterisk) with an aspartate and glutamate at the +2 and +3 positions after the acylated cysteine (underlined). The soluble periplasmic variant (YraPperi) is fused to the DsbA signal peptide (purple) that is cleaved upon export to the periplasm via the Sec system. (B-I) Overnight cultures of (B) TB140 (ΔenvC) or MT135 (ΔenvC ΔyraP) either (C) alone or harboring the integrated construct (D) attλMT196 (Plac::yraPWT), (E) attλMT198 (Plac::yraPIM), (F) attλMT209 (Plac::ssdsbA-yraPperi), (G) attλMT197 (Plac::yraPWT-mCherry), (H) attλMT199 (Plac::yraPIM-mCherry), or (I) attλMT210 (Plac::ssdsbA-yraPperi-mCherry) were diluted in minimal M9-maltose medium only (B-C) or supplemented with 10μM (D, F-G, I) or 20μM (E, H) IPTG. Cells were grown at 30°C to an OD600 of 0.2–0.25 before they were visualized on 2% agarose pads with DIC optics. Bar = 10μm.