Abstract
Blue nevi are melanocytic tumors originating in the cutaneous dermis. Malignant tumors may arise in association with or resembling blue nevi, so called ‘blue nevus-like melanoma’, which can metastasize and result in patient death. Identifying which tumors will behave in a clinically aggressive manner can be challenging. Identifying genetic alterations in such tumors may assist in their diagnosis and prognostication. Blue nevi are known to be genetically related to uveal melanomas (eg, both harboring GNAQ and GNA11 mutations). In this study, we analyzed a large cohort (n=301) of various morphologic variants of blue nevi and related tumors including tumors diagnosed as atypical blue nevi (n=21), and blue nevus-like melanoma (n=12), screening for all gene mutations known to occur in uveal melanoma. Similar to published reports, we found the majority of blue nevi harbored activating mutations in GNAQ (53%) or GNA11 (15%). In addition, rare CYSLTR2 (1%) and PLCB4 (1%) mutations were identified. EIF1AX, SF3B1, and BAP1 mutations were also detected, with BAP1 and SF3B1 R625 mutations being present only in clearly malignant tumors (17% (n=2) and 25% (n =3) of blue nevus-like melanoma, respectively). In sequencing data from a larger cohort of cutaneous melanomas, this genetic profile was also identified in tumors not originally diagnosed as blue nevus-like melanoma. Our findings suggest that the genetic profile of coexistent GNAQ or GNA11 mutations with BAP1 or SF3B1 mutations can aid the histopathological diagnosis of blue nevus-like melanoma and distinguish blue nevus-like melanoma from conventional epidermal-derived melanomas. Future studies will need to further elucidate the prognostic implications and appropriate clinical management for patients with tumors harboring these mutation profiles.
Blue nevi are common dermal melanocytic proliferations of the skin.1 They derive their name from the blue color observed when viewed clinically, an appearance attributed to the Tyndall effect. The latter describes the phenomenon whereby longer wave lengths of light penetrate more deeply in the skin and are absorbed by deep dermal lying melanin pigment, whereas blue light, having a shorter wave length, is reflected closer to the skin surface.2 The most frequent blue nevus subtype is the common or Jadasohn–Tièche blue nevus.3 A range of other less frequent subtypes have also been described including cellular, epithelioid, sclerotic, and plaque-type blue nevi.1,2,4
The overwhelming majority of blue nevi are benign and are often only biopsied to rule out a primary melanoma or a metastasis. Rare malignant tumors showing some of the morphologic features seen in blue nevi are often termed ‘malignant blue nevi’ or ‘blue nevus-like melanoma’. The latter term is preferable because the term ‘malignant blue nevus’ includes the contradictory terms ‘malignant’ and ‘nevus’ (which by definition connotes a benign melanocytic proliferation). Blue nevus-like melanomas are indeed malignant tumors and should be treated accordingly.5–8 If tumors resembling blue nevi show atypical clinico-pathologic features that are considered to fall short of malignancy, and there is consequently uncertainty with regard to their likely biological behavior, they are often designated ‘atypical cellular blue nevi’ (denoting a tumor of uncertain or indeterminate biologic potential). Unfortunately, no consensus histological criteria for atypical cellular blue nevi exist and diagnostic interobserver reproducibility is poor,9 however, features proposed to render this diagnosis include one or more of the following features: asymmetry, hypercellular foci, focal cytological atypia, and mitoses (depending on the literature < 2 or < 3 per mm2).1,2,7,8 Necrosis and atypical mitotic features are usually considered diagnostic of melanoma (blue nevus-like melanoma).
Distinguishing blue nevus-like melanoma from blue nevi histologically can be challenging. Many conventional histological criteria established to identify malignant behavior in more frequently occurring epidermal-derived melanomas cannot be applied to blue nevi. For example, blue nevi do not show maturation of cell nests toward deeper parts of the tumor, nor do malignant proliferations demonstrate ascending melanocytes in the epidermis (ie, pagetoid epidermal invasion), both useful diagnostic features frequently observed in epidermal-originating melanomas. Blue nevi also uniformly express HMB-45, an immunohistochemical marker which in epidermal-derived melanocytic tumors is usually only present in superficial melanocytes and if expressed in deeper dermal raises the possibility of melanoma. These features can make the distinction between benign and malignant dermal melanocytic proliferations difficult.
Blue nevi are genetically distinct from epidermal-derived common nevi and melanoma. Epidermal-derived nevi and melanoma frequently harbor mutations in BRAF and NRAS.10–12 This is uncommon in blue nevi which frequently contain activating GNAQ, GNA11 and less frequently CYSLTR2 mutation.13–16 All of these mutations were also identified in uveal melanoma.13–15 Other genes mutated in uveal melanomas are BAP1, EIF1AX, and SF3B1.17–19 Interestingly, mutations in these three genes are almost always mutually exclusive and have prognostic relevance. In uveal melanomas, BAP1 mutations resulting in loss of BAP1 protein function are associated with a poor prognosis,18 whereas SF3B1 and EIF1AX mutations are generally associated with tumors having a favorable prognosis.17,19
The genetic similarity between blue nevi and uveal melanoma has been further demonstrated by a number of recent studies reporting BAP1 mutations or protein loss occurring in blue nevus-like melanoma.5,20–23 Similar to uveal melanocytic tumors, this fits the classic progression model, where benign tumors (nevi) frequently harbor an initial mutation (ie, a GNAQ or GNA11 mutation) and acquisition of additional genetic alterations (ie, a SF3B1, EIF1AX or BAP1 mutation) results in progression to a malignant neoplasm (melanoma).
The goal of the current study was to determine the extent to which the genetic alterations identified in uveal melanoma are also present in blue nevi, atypical cellular blue nevi, and blue nevus-like melanoma, and could potentially be applied to distinguish benign from malignant tumors. A targeted next-generation sequencing panel covering all known recurrent mutations in these genes in uveal melanoma was applied to a cohort of 301 blue nevi of various subtypes, including tumors diagnosed as atypical cellular blue nevi or blue nevus-like melanoma.
Materials and methods
Sample Selection
Samples of blue nevi, atypical cellular blue nevi and blue nevus-like melanoma were obtained by searching the databases of the Melanoma Institute Australia (n=26), the Department of Dermatology University Hospital Essen (n=72), Dermatopathologie Friedrichshafen (n=106), Pathologie Salzburg (n =32) and Dermatopathologie bei Mainz (n=65), Germany. All cases were assessed histologically by at least two board-certified pathologists or dermatopathologists (RS, LJ, UH, RM, ME, AR, HK, HM and KGG). Of these samples, approximately one third (103) of the cases were described in a previous publication.16 The study was performed in accordance with the guidelines of the ethics committee of the University of Duisburg-Essen and approved under the IRB-number 16–6951-BO.
DNA Isolation
DNA was isolated from formalin fixed paraffin-embedded tumor tissue cut in 10 m-thick sections. Sections were then deparaffinized and manually macrodissected according to standard procedures. Genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions.
Targeted Sequencing
A custom amplicon-based sequencing panel covering ten genes (Table 1) was designed. Included were all seven genes known to occur in blue nevi and uveal melanoma (GNAQ, GNA11, PLCB4, CYSLTR2, EIF1AX, SF3B1 and BAP1) as well as the most frequent activating gene mutations in cutaneous nevi and cutaneous melanomas (BRAF, NRAS and KIT). Coverage focused on the known relevant mutational hotspots for all genes with the exception of BAP1, where relevant inactivating mutations have been reported throughout the gene.18
Table 1.
Genes covered in the applied sequencing panel
| No. | Gene | Chr. | Location GRCh37 | Target exons | Selection of mutations covered | Mutation type | Primer pairs |
|---|---|---|---|---|---|---|---|
| 1 | BRAF | 7 | 140453065 | 11, 15 | G463, G465, V600 | Activating | 4 |
| 2 | NRAS | 1 | 115256411 | 1, 2 | G12, G13, Q61 | Activating | 5 |
| 3 | KIT | 4 | 55593572 | 11, 13, 17 | L576, K642, N822 | Activating | 8 |
| 4 | GNAQ | 9 | 80409369 | 4, 5 | R183, Q209 | Activating | 6 |
| 5 | GNA11 | 19 | 3114932 | 4, 5 | R183, Q209 | Activating | 3 |
| 6 | CYSLTR2 | 13 | 49281314 | 1 | L129 | Activating | 1 |
| 7 | PLCB4 | 20 | 9389740 | 20 | D630 | Activating | 1 |
| 8 | SF3B1 | 2 | 198267458 | 14 | R625 | Change of function | 1 |
| 9 | EIF1AX | X | 20156647 | 1, 2 | Mutations in exons 1 and 2 | Change of function | 3 |
| 10 | BAP1 | 3 | 52436293 | all | Mutations in all exons | Inactivating | 41 |
Panel design and preparation took place applying the GeneRead Library Prep Kit from QIAGEN according to the manufacturer’s instructions. NEB-Next Ultra DNA Library Prep Mastermix Set and NEB-Next Multiplex Oligos for Illumina from New England Biolabs were applied for adapter ligation and barcoding of individual samples. Sequencing was performed on an Illumina MiSeq next-generation sequencer, with up to 70 samples sequenced in parallel. Sequencing the sample cohort, an average coverage of 5753 reads was achieved with 495% of the target area having a minimum coverage of 30 reads.
Sequence Analysis
CLC Cancer Research Workbench from QIAGEN was applied for sequence analysis, applying a number of steps as previously reported24 and demonstrated here briefly. The CLC workflow included adapter trimming and read pair merging before mapping to the human reference genome (hg19). Subsequently insertions and deletions as well as single nucleotide variant detection, local realignment, and primer trimming followed. Various databases (COSMIC, ClinVar, dbSNP, 1000 Genomes Project, HAPMAP, and PhastCons-Conservation_scores_hg19) were cross-referenced to obtain additional information regarding mutation type, single nucleotide polymorphisms, and conservation scores. Resulting csv files were further analyzed manually screening for mutations affecting the protein coding portion of the gene predicted to result in non-synonymous amino acid changes. Mutations were considered if the overall coverage of the mutation site was ≥30 reads, ≥5 reads reported the mutated variant and the frequency of mutated reads was ≥3%.
BAP1 Immunohistochemistry
BAP1 immunohistochemistry was performed applying a rabbit polyclonal antibody recognizing amino acids 430–729 of the BAP1 protein (clone C-4, Santa Cruz Biotechnology) as previously reported.25 Nuclear staining was assessed for positivity.
Genetic Screening for Tumors with a Blue Nevus-Like Melanoma Mutation Profile
The database of sequenced melanoma samples (n=1121) in Essen was screened for tumors where an activating GNAQ or GNA11 hotspot mutation had been identified. Tumors harboring these mutations, not reported to be of uveal or central nervous system origin, were assessed in more detail, screening available clinical data. If a cutaneous origin was reported, archived isolated tumor DNA was selected and sequenced applying the aforementioned 10 gene panel (Table 1). Particular attention was given to potential BAP1, SF3B1 and EIF1AX mutations. When tissue block material was still available, BAP1 immunohistochemistry was also performed.
Associations of Gene Mutation Status with Clinical and Pathologic Parameters
We investigated associations of mutation status with available clinical and pathological parameters using Kruskal–Wallis tests, χ2 tests and Fisher exact tests as appropriate. All statistical analyses were performed using IBM SPSS Statistics software (version 20.0; International Business Machines, Armonk, NY, USA). A P-value of ≤0.05 was considered statistically significant.
Results
Sample Cohort
The study cohort consisted of 301 blue nevus samples from 301 patients (174 females and 119 males, 8 cases unknown) with an average age of 48 years (range 3–99). The tumors included 176 (59%) common blue nevi, 69 (23%) cellular blue nevi, 21 (7%) atypical cellular blue nevi and 12 (4%) blue nevus-like melanoma. In addition, 18 (6%) combined nevi, harboring a blue nevus component, were assessed and 5 (2%) tumors were analyzed where genetic data suggested a blue nevus-like melanoma. All samples assessed were primary tumors, with the exception of samples suggested to be blue nevus-like melanoma by genetic analysis (5 tumor samples, including 4 metastases and 1 primary), defined as melanomas of cutaneous origin where sequencing identified activating GNAQ or GNA11 mutations. An overview of available clinical data is listed in Table 2.
Table 2.
Blue nevi cohort, clinical and genetic overview
| Histological blue nevus subtype | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||||
| Common BN |
Cellular BN |
Combined nevus |
Atypical cellular BN |
BNLM
|
Genetically BNLM |
Total
|
|||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | ||
| Total | 176 | 56 | 69 | 23 | 18 | 6 | 21 | 7 | 12 | 4 | 5 | 2 | 301 | ||
| Age | |||||||||||||||
| Median | P < 0.001 | 53 | 35 | 42 | 29 | 55 | 39 | 48 | |||||||
| Range | 7–99 | 8–82 | 13–67 | 3–79 | 23–88 | 20–66 | 3–99 | ||||||||
| Sex | |||||||||||||||
| Female | P = 0.11 | 101 | 57 | 45 | 65 | 12 | 67 | 9 | 43 | 5 | 42 | 2 | 40 | 174 | 58 |
| Male | 73 | 42 | 20 | 29 | 6 | 33 | 10 | 48 | 7 | 58 | 3 | 60 | 119 | 40 | |
| Missing data | 2 | 1 | 4 | 6 | 0 | 0 | 2 | 10 | 0 | 0 | 0 | 0 | 8 | 3 | |
| Site | |||||||||||||||
| Head/neck | P = 0.29 | 47 | 27 | 15 | 22 | 6 | 33 | 5 | 24 | 5 | 42 | 3 | 60 | 81 | 27 |
| Trunk | 42 | 24 | 27 | 39 | 6 | 33 | 7 | 33 | 3 | 25 | 2 | 40 | 87 | 29 | |
| Upper extremity | 31 | 18 | 8 | 12 | 0 | 0 | 2 | 10 | 0 | 0 | 0 | 0 | 41 | 14 | |
| Lower extremity | 26 | 15 | 10 | 15 | 4 | 22 | 3 | 14 | 0 | 0 | 0 | 0 | 43 | 14 | |
| Missing data | 30 | 17 | 9 | 13 | 2 | 11 | 4 | 19 | 4 | 33 | 0 | 0 | 49 | 16 | |
| Activating mutations | |||||||||||||||
| GNAQ | P < 0.001 | 91 | 52 | 50 | 73 | 0 | 0 | 14 | 67 | 3 | 25 | 2 | 40 | 160 | 53 |
| GNA11 | 30 | 17 | 8 | 12 | 0 | 0 | 0 | 0 | 4 | 33 | 3 | 60 | 45 | 15 | |
| CYSLTR2 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 4 | 1 | |
| PLCB4 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 3 | 1 | |
| BRAF | 0 | 0 | 1 | 1 | 9 | 50 | 2 | 10 | 3 | 25 | 0 | 0 | 15 | 5 | |
| NRAS | 3 | 2 | 0 | 0 | 4 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 2 | |
| Wild-type | 48 | 27 | 10 | 15 | 5 | 28 | 3 | 14 | 2 | 17 | 0 | 0 | 68 | 23 | |
Abbreviations: BN, blue nevus; BNLM, blue nevus-like melanoma.
Distribution of Activating Oncogene Driver Mutations
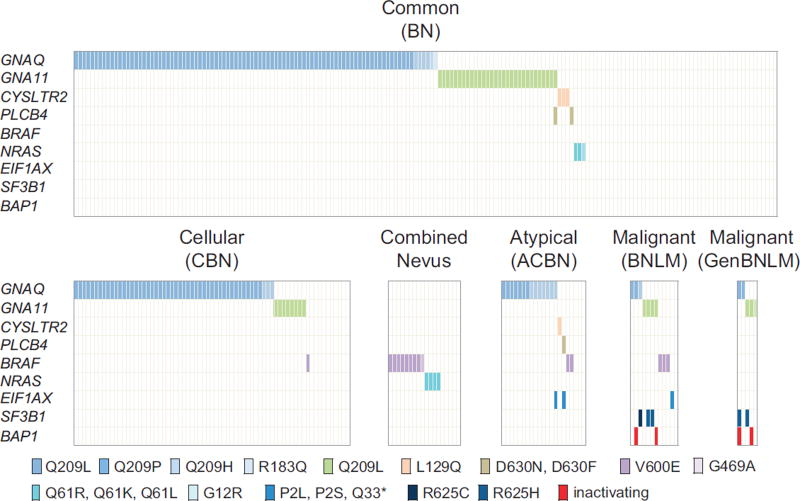
Targeted amplicon sequencing of all genes, as described in the Material and methods section, was performed for the entire tumor cohort. Consistent with previously reported results, the most frequent activating mutations identified were in GNAQ, 53% (143 c.626A>T Q209L, 15 c.626A>C Q209P, 1 c.627A>T A209H and 1 c.548G>A R183Q) and GNA11, 15% (44 c.626A>T Q209L and 1 c.547C>T R183C) (Figure 1). Fifteen (5%) BRAF mutations (12 c.1799A>T V600E and 1 c.227G>C G469A) and 7 (2%) NRAS mutations (3 c.182A>G Q61R, 3 c.181C>A Q61K and 1 c.34G>C G12R) were detected, most of these occurring in the combined nevi group (and was likely derived from the non-blue nevus component of the lesion). Three (1%) PLCB4 mutations (2 c.1888G>A D630N and 1 c.1888_1889delGAinsTT D630F) were identified. Four tumors (1%) harbored activating CYSLTR2 mutations (c.386T>A L129Q). Generally, activating mutations were found to be mutually exclusive, with the exception of a GNA11 Q209L and PLCB4 D630N occurring mutually in one common blue nevus sample. The mutations and distribution identified are further shown in Figure 1.
Figure 1.
Mutations identified in a cohort of blue nevi and blue nevus-like melanomas. Shown are the activating mutations identified by targeted next-generation sequencing in a cohort of 301 blue nevi. The protein alterations occurring through the different mutations are signified by different colors, annotated at the bottom of the figure. ACBN, atypical cellular blue nevus; BNLM, blue nevus-like melanoma; BN, blue nevus; CBN, cellular blue nevus; combined nevus, nevus having a blue and conventional nevus compartment; GenBNLM, genetic BNLM.
Distribution of Mutations in Combined Nevi
In combined nevi, which demonstrated a clear conventional epidermal or compound (epidermal and dermal) nevus component adjacent to or admixed with a blue nevus component, the mutations identified were primarily BRAF mutations (50%, 8 V600E and 1 G469A) and NRAS mutations (22%, 2 Q61R and 2 Q61K).
Distribution of Mutations in Atypical Cellular Blue Nevi and Blue Nevus-Like Melanoma
Our cohort included 21 tumors diagnosed as atypical cellular blue nevi and 12 tumors diagnosed as blue nevus-like melanoma (Table 3). In the atypical cellular blue nevi, 66% (n= 14) of tumors harbored activating GNAQ mutations. One (5%) CLYSTR2 and one (5%) PLCB4 mutation (Supplementary Figure 1) were identified. Two tumors (10%) were found to harbor BRAF V600E mutations. Two mutations in EIF1AX (exon 1 or 2) were noted, a P2L and Q33* mutation, respectively (Table 3 and Supplementary Figure 1).
Table 3.
Genetic alterations in atypical blue nevi and blue nevus-like melanoma
| Nr | Type ACBN |
More detailed diagnosis given | Age: | Sex: | Location | BRAF 600 |
NRAS 12,13,61 |
GNA11 183,209 |
GNAQ 183,209 |
CYSLTR2 129 |
PLCB4 630 |
EIF1AX Ex.1+2 |
SF3B1 625 |
BAP1 inact. |
BAP1 IHC |
Status | Int. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ACBN | atypical pigmented cellular blue nevus | 47 | F | upper chest | - | - | - | - | - | - | - | - | - | + | N.K. | - |
| 2 | ACBN | animal type blue nevus | N.K. | N.K. | N.K. | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 3 | ACBN | atypical cellular blue nevus | 79 | M | right neck | - | - | - | Q209P | - | - | - | - | - | + | N.K. | - |
| 4 | ACBN | atypical cellular blue nevus | 42 | M | left occipital | - | - | - | Q209P | - | - | - | - | - | + | F | 1 |
| 5 | ACBN | atypical cellular blue nevus | 23 | F | right hand | - | - | - | Q209P | - | - | - | - | - | N.A. | N.K. | - |
| 6 | ACBN | atypical cellular blue nevus | 3 | F | scalp | - | - | - | Q209P | - | - | - | - | - | + | N.K. | - |
| 7 | ACBN | atypical cellular blue nevus | 44 | M | right thigh | - | - | - | - | - | D630F | P2L | - | - | + | F | 13y |
| 8 | ACBN | atypical cellular blue nevus | 66 | M | N.K. | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 9 | ACBN | atypical cellular blue nevus | 29 | M | N.K. | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 10 | ACBN | atypical neuronevus Masson | 26 | M | gluteal left | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 11 | ACBN | atypical neuronevus Masson | 35 | W | gluteal left | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 12 | ACBN | atypical epithelold blue nevus | N.K. | N.K. | N.K. | - | - | - | - | - | - | - | - | - | + | N.K. | - |
| 13 | ACBN | pigmented epithelioid melanocytoma | 17 | M | right jaw | - | - | - | - | L129Q | - | - | - | - | + | F | 1y |
| 14 | ACBN | pigmented epithelioid melanocytoma | 17 | F | right back | - | - | - | Q209P | - | - | - | - | - | + | F | 6y |
| 15 | ACBN | pigmented epithelioid melanocytoma | 26 | F | right cheek | - | - | - | Q209P | - | - | Q33* | - | - | + | F | 2y |
| 16 | ACBN | atypical neuronevus Masson | 28 | M | gluteal left | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 17 | ACBN | atypical neuronevus Masson | 66 | F | gluteal | - | - | - | Q209P | - | - | - | - | - | + | N.K. | - |
| 18 | ACBN | atypical spindle cell dermal melanocyte tumor | 63 | M | back of left foot | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 19 | ACBN | atypical epithelioid dermal melanocyte tumor | 29 | M. | upper back | - | - | - | - | - | - | - | - | - | + | N.K. | - |
| 20 | ACBN | atypical cellular blue nevus | 46 | F | upper arm right | V600E | - | - | - | - | - | - | - | - | N.A. | ||
| 21 | ACBN | atypical cellular blue nevus | 28 | F | lower right calf | V600E | - | - | - | - | - | - | - | - | + | F | 4y |
|
| |||||||||||||||||
| BNLM | Diagnosis given | Age | Sex | Location | BRAF | NRAS | GNA11 | GNAQ | CYSLTR2 | PLCB4 | EIF1AX | SF3B1 | BAP1 | BAP1 | Status | Int. | |
|
| |||||||||||||||||
| 22 | BNLM | blue nevus-like melanoma | 88 | M | posterior left scalp | - | - | - | - | - | - | P2S | - | - | + | † | * |
| 23 | BNLM | blue nevus-like melanoma | 61 | M | scalp | - | - | Q209L | - | - | - | - | R625H | - | + | † | * |
| 24 | BNLM | blue nevus-like melanoma | 70 | M | left back | - | - | - | Q209P | - | - | - | R625C | - | N.K.# | † | * |
| 25 | BNLM | blue nevus-like melanoma | 63 | F | head right side | - | - | Q209L | - | - | - | - | - | - | N.A. | F | * |
| 26 | BNLM | blue nevus-like melanoma | 35 | M | gluteal region left | - | - | - | Q209L | - | - | - | - | - | + | N.K. | - |
| 27 | BNLM | blue nevus-like melanoma | 39 | M | scrotum left | - | - | Q209L | - | - | - | - | R625H | - | + | A | * |
| 28 | BNLM | blue nevus-like melanoma | 61 | F | N.K. | - | - | - | - | - | - | - | - | - | + | N.K. | - |
| 29 | BNLM | blue nevus-like melanoma | 28 | F | N.K. | - | - | Q209L | - | - | - | - | - | P190R | neg | N.K. | - |
| 30 | BNLM | blue nevus-like melanoma | 49 | F | occipital | - | - | - | Q209L | - | - | - | - | A95fs | neg | N.K. | - |
| 31 | BNLM | blue nevus-like melanoma | 51 | M | buttock | V600E | - | - | - | - | - | - | - | - | N.A. | N.K. | - |
| 32 | BNLM | blue nevus-like melanoma | 84 | F | N.K. | V600E | - | - | - | - | - | - | - | - | + | N.K. | - |
| 33 | BNLM | blue nevus-like melanoma | 23 | M | scalp | V600E | - | - | - | - | - | - | - | - | + | N.K. | - |
|
| |||||||||||||||||
| Gen BNLM | Clinical Information | Age | Sex | Location | BRAF | NRAS | GNA11 | GNAQ | CY5LTR2 | PLCB4 | EIF1AX | SF3B1 | BAP1 | BAP1 | Status | Int. | |
|
| |||||||||||||||||
| 34 | GenBNLM | MUP - "metastasis" left axilla | 39 | F | axillary left | - | - | - | Q209L | - | - | - | R625C | P175F | neg | N.K. | - |
| 35 | GenBNLM | melanoma NOCTT >4 mm | 20 | W | gluteal right | - | - | - | Q209L | - | - | - | - | - | N.A. | F | 3y |
| 36 | GenBNLM | melanoma NOC TT 7.5 mm | 64 | M | temporal right | - | - | Q209L | - | - | - | - | R625C | - | + | A | * |
| 37 | GenBNLM | superficial spreading melanoma | 66 | M | pectoral | - | - | R183C | - | - | - | - | - | - | + | F | 1y |
| 38 | GenBNLM | superficial spreading melanoma | 31 | M | temporal right | - | - | Q209L | - | - | - | - | - | Q253* | neg | A | * |
Abbreviations: A, alive; ACBN, atypical cellular blue nevus; blue, mutations altering function; BNLM, blue nevus-like melanoma (also known as ‘malignant blue nevus’); F, free of disease; GenBNLM, genetic blue nevus-like melanoma; Green, activating mutations; IHC, immunohistochemistry; Int., interval (available follow-up period); Max., maximum; Met., metastasis; MUP, melanoma of unknown primary; N.A., not analyzed; N.K., not known; NOC, not otherwise characterized; red, loss of function mutations; TT, tumor thickness.
Signifies cases where more detailed follow-up information.
BAP1 immunohistochemistry was inconclusive (demonstrated in Supplementary Figure 2).
The 12 tumors diagnosed as blue nevus-like melanoma harbored mutations in GNAQ in 25% (n=3), GNA11 in 33% (n=4), and BRAF in 25% (n= 3) of cases (Figures 2, 3, 4 and Supplementary Figures 2). One tumor (8%) contained an EIF1AX P2L mutation. Three tumors (25%) harbored SF3B1 R625 mutations (2 R625H and 1 R625C mutation, Figures 2, 3, Supplementary Figure 2). Two inactivating BAP1 mutations were identified, with loss of protein confirmed by immunohistochemistry (Figure 4 and Supplementary Figure 3). All tumors demonstrating SF3B1 or BAP1 mutations additionally harbored activating GNAQ or GNA11 mutations. In two tumors, the SF3B1 (Figure 3) and BAP1 (Figure 4) mutations were detected in the melanoma, but absent in the nevus portion of the tumor.
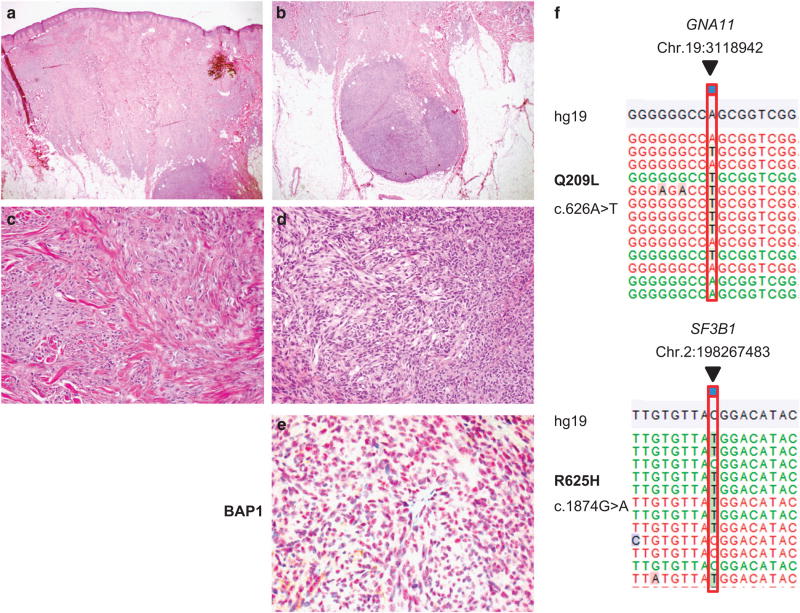
Figure 2.
Blue nevus-like melanoma with a GNA11 and SF3B1 mutation. Demonstrating a case of a tumor histopathologically diagnosed as a blue nevus-like melanoma. In addition to a larger dermal component (a, c × 20), a nodule of cells is demonstrated in the subcutis (b, d ×200). The BAP1 immunohistochemistry (e ×400) showed retained BAP1 protein expression. (f) The GNA11 Q209L (c.626A>T) and SF3B1 R625H (c.1874G>A) mutations identified in the tumor. Annotation according to human genome assembly 19 (hg19). (The case demonstrated refers to number 23 in Table 3).
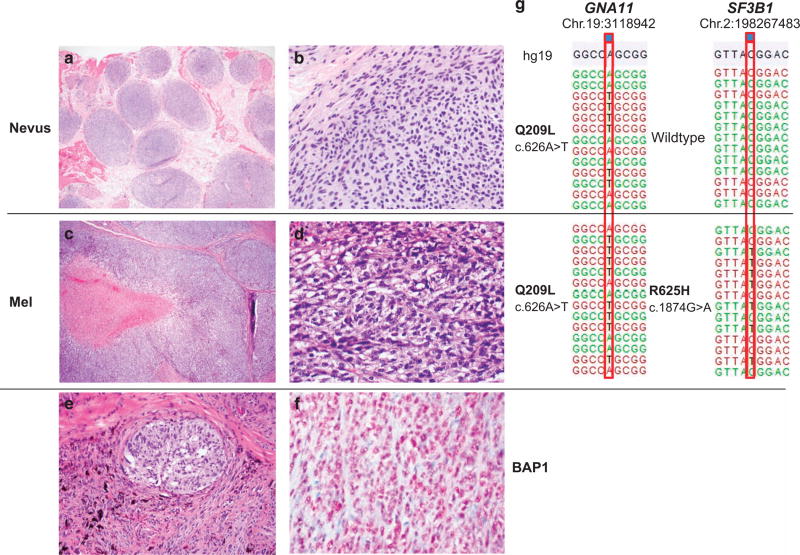
Figure 3.
Cellular blue nevus progressing to melanoma by SF3B1 mutation. A tumor arising in the scrotum of a 39-year-old male. Upon initial excision, a melanoma was diagnosed (c × 20 and d × 400), which demonstrated both highly pleomorphic cells and areas of necrosis (c). Near the surgical margins, a benign dermal melanocytic proliferation was present with features of a cellular blue nevus (a × 40 and b × 400). In e (×200), mainly benign blue nevus cells are present with a focal nodule of malignant cells infiltrating in the center of the field of view. The BAP1 immunohistochemistry showed retained BAP1 protein expression both in the benign and malignant portions of the lesion (f × 400). Genetic results are shown in g. The GNA11 Q209L (c.626A>T) mutation was detected in both the nevus and melanoma portions of the tumor however, the SF3B1 R625H (c.1874G>A) mutation was only identified in the melanoma. Annotation according to human genome assembly 19 (hg19). (The case demonstrated refers to number 27 in Table 3). Mel, melanoma.
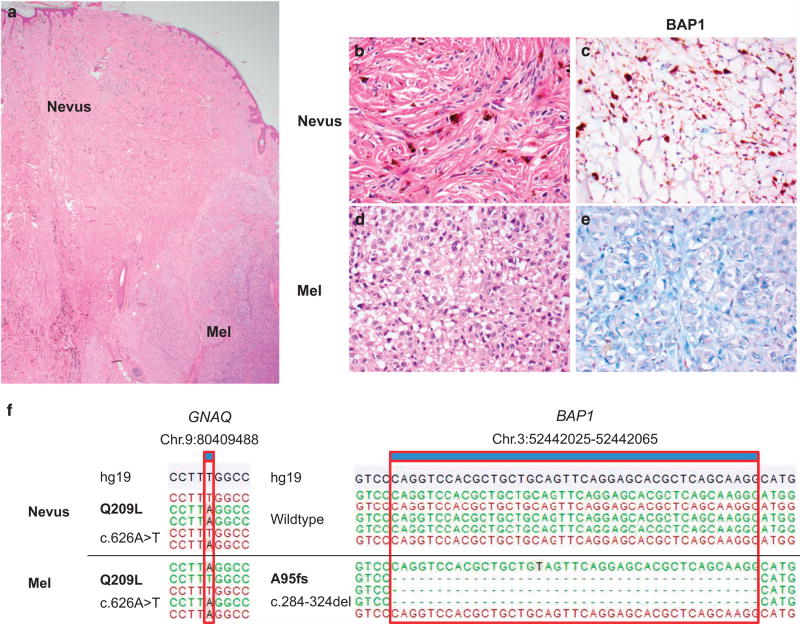
Figure 4.
Blue nevus progressing to melanoma by inactivating BAP1 mutation. A case demonstrating a residual blue nevus proliferation (a × 20) in the top left and progression to melanoma in the bottom right. High magnification of the nevus and melanoma portions of the lesion with BAP1 immunohistochemistry is demonstrated in b–e ×400. The top panel demonstrated morphologically benign pigmented nevus cells with the corresponding BAP1 expression in immunohistochemistry. The bottom panel demonstrates the melanoma fraction with epithelioid cells and lack of BAP1 expression (right). (c) The genetic data, demonstrating the same GNAQ Q209L (c.626A>T) mutation in both the nevus and melanoma portions of the tumor. On the right, the inactivating BAP1 mutation leading to a frame-shift mutation (A95fs, c.284-324del) was only found in the melanoma, not the nevus portion of the tumor. The notation is according to human genome assembly 19 (hg19). (The case presented is number 30 in Table 3). Mel, melanoma.
Tumors With a Blue Nevus-Like Melanoma Mutation Profile
By screening existing mutation data for melanomas harboring GNAQ or GNA11 mutations not reported as tumors of uveal or CNS origin, we identified five tumors. Three of these were tumors demonstrated unusual or atypical clinical or histopathological features compared with conventional melanomas. In one case, the tumor was diagnosed as a MUP (melanoma of unknown primary) and presented as a subcutaneous tumor in the axilla. Genetic screening of the tumor revealed mutations in GNA11, SF3B1, and BAP1 (case 34, Table 3 and Figure 5). In two other cases, the melanoma subtype was not documented in the original pathology and the original pathology slides were not available for review; both tumors were thick melanomas (>4 mm and >7.5 mm, case 35 and case 36 in Table 3, respectively). In the remaining two cases, the melanoma subtype was reported as superficial spreading melanoma, suggesting the primary tumor had a junctional melanocytic component, a feature not usually present in blue nevi (or blue nevus-like melanoma). One of these two tumors (case 38, Table 3) also had a clear inactivating BAP1 mutation, confirmed by loss of BAP1 expression with immunohistochemistry (Supplementary Figure 5) further fitting the genetic profile of a blue nevus-like melanoma.
Figure 5.
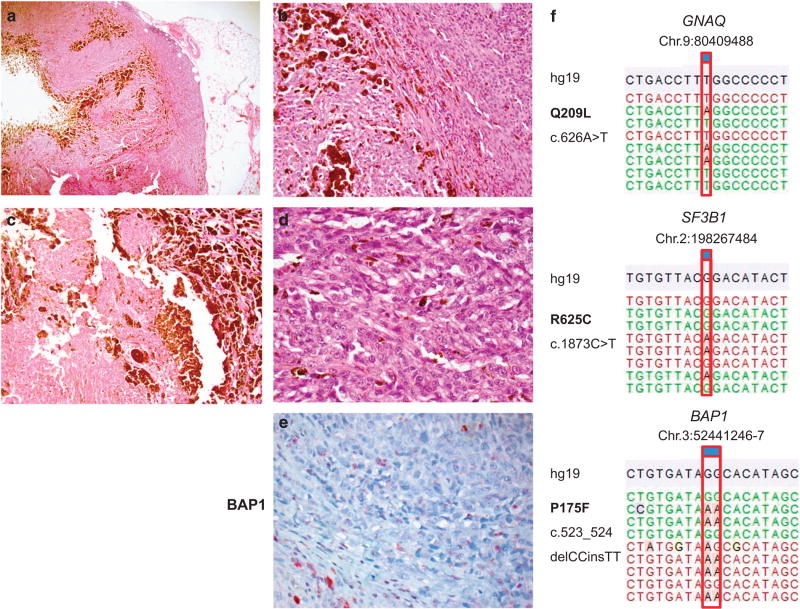
Metastasized melanoma harboring a GNAQ, SF3B1, and BAP1 mutation. Example of a genetically diagnosed blue nevus-like melanoma. The tumor was originally diagnosed as a melanoma of unknown primary (MUP). The morphology demonstrated in a ×40, b ×100, c ×200, and d ×400 shows heavy pigmentation, reminiscent of a blue nevus-like melanocytic tumor, with cytological features of malignancy. BAP1 expression was lost in the tumor by immunohistochemistry (e ×400). Interestingly, this tumor showed presence of mutations in GNAQ Q209L (c.626A>T), SF3B1 R625C (c.1873C>T), and BAP1 P175F (c.523_524delCCinsTT). Annotation according to human genome assembly 19 (hg19). (The case presented is number 34 in Table 3).
Associations of Clinical and Pathological Parameters with Oncogene Mutation Status
Statistically significant associations of melanoma subtype with patient age and with mutation type were identified; full details are presented in Table 2. In all cases of blue nevus-like melanoma where follow-up data was available, this was screened and the relevant information is presented in Table 4. Associations with survival were not performed as the available data was too limited to allow a meaningful analysis.
Table 4.
Available clinical information of malignant tumors
| No. | Type | More detailed diagnosis given | Gene mutations | Clinical info |
|---|---|---|---|---|
| 22 | BNLM | Blue nevus-like melanoma | EIF1AX | Multiple local/in-transit recurrence 16, 18, 23, 26, and 28 months after diagnosis. Patient died 30 months after diagnosis (Alzheimer’s disease, heart failure, and metastasized melanoma). |
| 23 | BNLM | Blue nevus-like melanoma | GNA11 SF3B1 | The patient had two other primary melanomas, one diagnosed 12 months prior to the BNLM, another nodular melanoma diagnosed ~48 months (4 years) later which had a positive SLNB. Patient died of metastatic disease ~ 72 months (6 years) after diagnosis. |
| 24 | BNLM | Blue nevus-like melanoma | GNAQ SF3B1 | Developed liver metastases and died of disease within 12 months. |
| 25 | BNLM | Blue nevus-like melanoma | GNA11 | Diagnosed with a non-ulcerated 7 mm tumor, shortly afterwards full tumor excision with wide excision margin and SLNB neg., no further disease to date (11 months follow-up). |
| 26 | BNLM | Blue nevus-like melanoma | GNAQ | Diagnosed with a tumor thickness >6mm (pT4b), SLNB pos. (1/1 LN), LAD neg. (0/10), no further disease to date (13 months follow-up). |
| 27 | BNLM | Blue nevus-like melanoma | GNA11 SF3B1 | Presented with an unusual tumor of the scrotum. Diagnostically a metastasis was considered, however upon histological and genetic data the diagnosis of a malignant blue nevus was made. Currently free of disease (4 months follow-up). |
| 35 | GenBNLM | Melanoma NOC TD > 4 mm | GNAQ | SLNB (2/2 pos.), LAD neg. (0/9), irregular follow-up, last seen 34 months after diagnosis with no sign of disease in staging. |
| 36 | GenBNLM | Melanoma NOC TD 7.5 mm | GNA11, SF3B1 | Cervical LN metastasis ~144 months (12 years) after diagnosis. Sequence analysis led to the genetic diagnosis of a blue nevus-like melanoma. Subsequently multiple additional LN and soft tissue metastasis in the neck area have been excised (~146–164 months after diagnosis). |
| 37 | GenBNLM | Superficial spreading melanoma TT 1.45 mm CL II | GNAQ | Diagnosed with a tumor thickness of 1.45 mm and Clark level II, excised with 1 cm safety margin, SLNB pos. (1/3 LN). |
| 38 | GenBNLM | Superficial spreading melanoma | GNA11, BAP1 | No tumor thickness given on initial diagnosis, local recurrence 25 months later, SLNB was performed and pos., LAD pos. (1/5), 42 months after diagnosis recurrence mastoid right, radical neck dissection. Fifty months after diagnosis liver metastasis, partial liver resection, Chemotherapy with gemcitabine and treosulfan, later ipilimumab, mixed response. Fifty-six months after diagnosis cutaneous nuchal and abdominal metastasis. Sixty-nine months after diagnosis scrotal metastasis. Seventy-seven months after diagnosis re-induction ipilimumab. Eighty-one months after diagnosis progressive disease. One-hundred three months after diagnosis genetic analysis identifying a genetic profile typical of a blue nevus-like melanoma. |
Abbreviations: BNLM, blue nevus-like melanoma; GenBNLM, genetic blue nevus-like melanoma; LAD, lymphadenectomy; neg, negative; NOC, not otherwise characterized; pos, positive; SLNB, sentinel lymph node biopsy; TT, tumor thickness.
Discussion
Our study presents the largest genetic screen of blue nevi and related tumors published to date. In addition to documenting the distribution of activating driver mutations in these tumors, it also further elucidates the unique genetic profile of malignant tumors, in particular highlighting the significance of the presence of BAP1 and SF3B1 R625 mutations in supporting a diagnosis of malignancy. This unique mutation profile may therefore have clinical implications, both from a diagnostic and treatment perspective.
Similar to previous studies,5,13,14,16 the overwhelming majority of blue nevi in our cohort were found to harbor activating mutations in GNAQ and GNA11. CYSLTR2 L129 and PLCB4 D630 mutations were rare with a frequency of around 1% (4 and 3 cases, respectively). BRAF and NRAS mutations were mostly observed in combined nevi (Figure 1) and rarely in tumors diagnosed as blue nevi. One can debate if tumors harboring BRAF and NRAS mutations should be considered blue nevi or not. For benign neoplasms, we would argue this question is purely academic and of no real clinical relevance. For atypical or malignant melanocytic proliferations, we believe that the term ‘blue nevus’ should be used with caution if a BRAF or NRAS mutation is detected. If this term is included in the diagnosis, additionally stating the mutation status may ensure that all available treatment options are considered (ie, BRAF inhibitor therapy).
Similar to uveal melanoma, our analysis found that EIF1AX, SF3B1, and BAP1 mutations also occur in blue nevus-like melanocytic proliferations. In our study, SF3B1 R625 and inactivating BAP1 mutations were only identified in tumors diagnosed as malignant (blue nevus-like melanoma). This is further supported by the available follow-up data. The BAP1 mutant tumor where follow-up data was available metastasized (case 38, Tables 3 and 4). In 5 tumors harboring SF3B1 mutations, follow-up data >1 year was available for 3 patients. In two cases, the patient died because of melanoma (case 23 and 24, Tables 3 and 4). Both cases had interesting clinical features. In case 24, the patient quickly died of liver metastasis, which in uveal melanoma typically occurs in patients whose tumor demonstrates BAP1 loss. Evidence for BAP1 alterations in case 24 was not observed in our study. Sequencing did not identify BAP1 mutations and immunohistochemistry was suboptimal and inconclusive (probably reflecting lack of antigen preservation because of the age of the tissue block, Supplementary Figure 1). The other patient (case 23) was diagnosed with two other primary melanomas. We were unable to definitively ascertain which tumor metastasized and led to the patient’s death. The last case for whom follow-up was available (case 36, Tables 3 and 4) definitely metastasized and showed no loss of BAP1 (by IHC and sequencing). This data is intriguing as in uveal melanomas SF3B1 mutations are associated with tumors which do not metastasize.17,19 Although larger cohorts will be required, the preliminary data we present here argues that both SF3B1 and BAP1 mutant tumors can metastasize and affected patients should be regularly followed up after primary complete excision.
For EIF1AX mutations (in exon 1 and 2), which occur in uveal melanomas and are associated with a favorable prognosis,17 their significance in blue nevus-like melanocytic proliferations is less clear. We identified EIF1AX mutations in two atypical cellular blue nevi and one blue nevus-like melanoma. On follow-up, the patients with atypical cases remained disease-free (Table 3), however the blue nevus-like melanoma recurred multiple times. Future studies will need to further elucidate the prognostic relevance of EIF1AX, SF3B1, and BAP1 mutations.
The cases of genetically defined blue nevus-like melanoma we present pose a number of unresolved questions. In cases originally diagnosed as a metastasis or melanoma not otherwise characterized (cases 34–36 in Tables 3, 4 and Figure 5), the clinical and histopathological description is retrospectively consistent with unrecognized blue nevus-like melanoma. Case 34 demonstrates how blue nevus-like melanoma may occasionally be misdiagnosed as a metastasis because of its deep location and absence of an associated epidermal component. In the other two cases (cases 37+38 in Tables 3 and 4) superficial spreading melanomas (SSM) were originally diagnosed implying epidermal tumor involvement, a feature not usually observed in blue nevus-like melanoma (unfortunately the original slides were not available for review). Potentially these cases represent melanomas arising from pre-existing combined nevi. Although a question of debate whether these tumors might actually be bona-fide epidermal-derived melanomas, we believe the unique genetic profile of such cases should be clearly communicated in diagnostic pathology reports (eg, ‘superficial spreading melanoma with the genetic profile of a blue nevus-like melanoma.’), as this could be relevant for patient management.
Our study has some limitations. Atypical cellular blue nevi and blue nevus-like melanoma are rare, which is why we could not avoid analyzing a heterogenous cohort from different institutions over different time periods. The original slides, clinical and follow-up data were not available in all cases, limiting the clinico-pathological associations that could be investigated. Our genetic analysis only covered the published relevant regions of 10 genes (Table 1) and paired normal DNA was not sequenced; as a result, rare mutations located outside the mutation hotspots or germline variants may not have been recognized. Our sequencing approach did not allow an assessment of copy number variations, which would have been valuable, as in particular chromosome 3 losses have been frequently reported in blue nevus-like melanomas.5,20,26
In our institutions, gene sequencing has become a routine part of the diagnostic evaluation of melanocytic tumors. If a tumor is histologically assessed as being clearly benign, no additional analysis is performed. However, in cases where the histopathologic diagnosis of an atypical cellular blue nevus or blue nevus-like melanoma is considered, IHC and genetic analysis are performed. Our standard melanoma sequencing panel covers all frequent activating mutations (BRAF, NRAS, KIT, NF1, GNAQ, GNA11, etc). If a GNAQ or GNA11 mutation is detected, the tumor is assumed to be a blue nevus-like melanocytic proliferation and sequencing of EIF1AX, SF3B1, and BAP1 is performed. If a clear SF3B1 R625 or inactivating BAP1 mutation (with IHC loss) is observed, we interpret this as molecular evidence in support of a diagnosis of blue nevus-like melanoma. The implications of an EIF1AX mutation are still unclear. Complete tumor excision and regular patient follow-up could be considered.
Owing to the morphological and genetic similarities a uveal melanoma metastasis should be excluded before diagnosing a blue nevus-like melanoma. In patients with no known history of uveal melanoma, a fundoscopic examination by an ophthalmologist is recommendable.
We believe our findings clearly demonstrate that in addition to known inactivating BAP1 mutations, SF3B1 R625 mutations in conjunction with GNAQ and GNA11 mutations are a genetic marker of malignancy in blue nevus-like melanocytic proliferations. This genetic profile can be applied as a diagnostic aid in cases where one is uncertain of a tumor’s malignant potential based on histopathologic assessment. In addition, the genetic profile may be used to diagnose blue nevus-like melanoma in melanomas where such a diagnosis was not suspected from histopathologic evaluation. Nevertheless, future studies will need to further assess the prognostic relevance of SF3B1 or BAP1 mutations in blue nevus-like melanoma.
Supplementary Material
Acknowledgments
The research was supported by a grant from the Deutsche Forschungsgemeinschaft (GR 3671/3-1) and the Deutsche Stiftung für Dermatologie (Stipendium zur Förderung der Dermatohistologie).
Dirk Schadendorf is on the advisory board or has received honararia from Roche, Genentech, Novartis, Amgen, GSK, BMS, Boehringer Ingelheim, and Merck. Bastian Schilling received honoraria from Roche, Bristol-Myers Squibb, and MSD Sharp & Dohme, research funding from Bristol-Myers Squibb and MSD Sharp & Dohme, and travel support from Roche, Bristol-Myers Squibb, and AMGEN. Elisabeth Livingstone received honoraria from Bristol-Myers Squibb, Boehringer-Ingelheim Pharma, Amgen, Roche, Novartis, MSD SHARP & DOHME, Merck and travel support from Amgen, Boehringer-Ingelheim Pharma, MSD SHARP & DOHME, and Novartis Pharma. Ioana Cosgarea has received travel support from Novartis, Bristol-Meyers Squibb, and TEVA.
Footnotes
Disclosure/conflict of interest
The remaining authors have nothing to declare.
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
References
- 1.Zembowicz A, Phadke PA. Blue nevi and variants: an update. Arch Pathol Lab Med. 2011:327–336. doi: 10.5858/2009-0733-RA.1. [DOI] [PubMed] [Google Scholar]
- 2.Zembowicz A, Mihm MC. Dermal dendritic melanocytic proliferations: an update. Histopathology. 2004:433–451. doi: 10.1111/j.1365-2559.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 3.Tièche M. Über benigne Melanome (“Chromatophorome”) der Haut - “blaue Neavi”. Virch Arch für path Anat. 1906:212–229. [Google Scholar]
- 4.Barnhill RL, Cerroni L, Cook M, et al. State of the art, nomenclature, and points of consensus and controversy concerning benign melanocytic lesions: outcome of an international workshop. Adv Anat Pathol. 2010:73–90. doi: 10.1097/PAP.0b013e3181cfe758. [DOI] [PubMed] [Google Scholar]
- 5.Costa S, Byrne M, Pissaloux D, et al. Melanomas associated with blue nevi or mimicking cellular blue nevi: clinical, pathologic, and molecular study of 11 cases displaying a high frequency of GNA11 mutations, BAP1 expression loss, and a predilection for the scalp. Am J Surg Pathol. 2016:368–377. doi: 10.1097/PAS.0000000000000568. [DOI] [PubMed] [Google Scholar]
- 6.Loghavi S, Curry JL, Torres-Cabala CA, et al. Melanoma arising in association with blue nevus: a clinical and pathologic study of 24 cases and comprehensive review of the literature. Mod Pathol. 2014:1468–1478. doi: 10.1038/modpathol.2014.62. [DOI] [PubMed] [Google Scholar]
- 7.Martin RC, Murali R, Scolyer RA, et al. So-called ‘malignant blue nevus’: a clinicopathologic study of 23 patients. Cancer. 2009:2949–2955. doi: 10.1002/cncr.24319. [DOI] [PubMed] [Google Scholar]
- 8.Murali R, McCarthy SW, Scolyer RA. Blue nevi and related lesions: a review highlighting atypical and newly described variants, distinguishing features and diagnostic pitfalls. Adv Anat Pathol. 2009:365–382. doi: 10.1097/PAP.0b013e3181bb6b53. [DOI] [PubMed] [Google Scholar]
- 9.Barnhill RL, Argenyi Z, Berwick M, et al. Atypical cellular blue nevi (cellular blue nevi with atypical features): lack of consensus for diagnosis and distinction from cellular blue nevi and malignant melanoma (‘malignant blue nevus’) Am J Surg Pathol. 2008:36–44. doi: 10.1097/PAS.0b013e3181573aaf. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shain AH, Yeh I, Kovalyshyn I, et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med. 2015:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 12.Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol. 2014:239–271. doi: 10.1146/annurev-pathol-012513-104658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 16.Moller I, Murali R, Muller H, et al. Activating cysteinyl leukotriene receptor 2 (CYSLTR2) mutations in blue nevi. Mod Pathol. 2016:350–356. doi: 10.1038/modpathol.2016.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harbour JW, Roberson ED, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh I, Mully TW, Wiesner T, et al. Ambiguous melanocytic tumors with loss of 3p21. Am J Surg Pathol. 2014:1088–1095. doi: 10.1097/PAS.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai J, Tetzlaff MT, Schuchter LM, Elder DE, Elenitsas R. Histopathologic and mutational analysis of a case of blue nevus-like melanoma. J Cutan Pathol. 2016:776–780. doi: 10.1111/cup.12731. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Alea M, Vivancos A, Caratu G, et al. Genetic profile of GNAQ-mutated blue melanocytic neoplasms reveals mutations in genes linked to genomic instability and the PI3K pathway. Oncotarget. 2016:28086–28095. doi: 10.18632/oncotarget.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivancos A, Caratu G, Matito J, et al. Genetic evolution of nevus of Ota reveals clonal heterogeneity acquiring BAP1 and TP53 mutations. Pigment Cell Melanoma Res. 2016:247–253. doi: 10.1111/pcmr.12452. [DOI] [PubMed] [Google Scholar]
- 24.van de Nes J, Gessi M, Sucker A, et al. Targeted next generation sequencing reveals unique mutation profile of primary melanocytic tumors of the central nervous system. J Neurooncol. 2016:435–444. doi: 10.1007/s11060-015-2052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Nes JA, Nelles J, Kreis S, et al. Comparing the prognostic value of BAP1 mutation pattern, chromosome 3 status, and BAP1 immunohistochemistry in uveal melanoma. Am J Surg Pathol. 2016:796–805. doi: 10.1097/PAS.0000000000000645. [DOI] [PubMed] [Google Scholar]
- 26.Held L, Eigentler TK, Metzler G, et al. Proliferative activity, chromosomal aberrations, and tumor-specific mutations in the differential diagnosis between blue nevi and melanoma. Am J Pathol. 2013:640–645. doi: 10.1016/j.ajpath.2012.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.