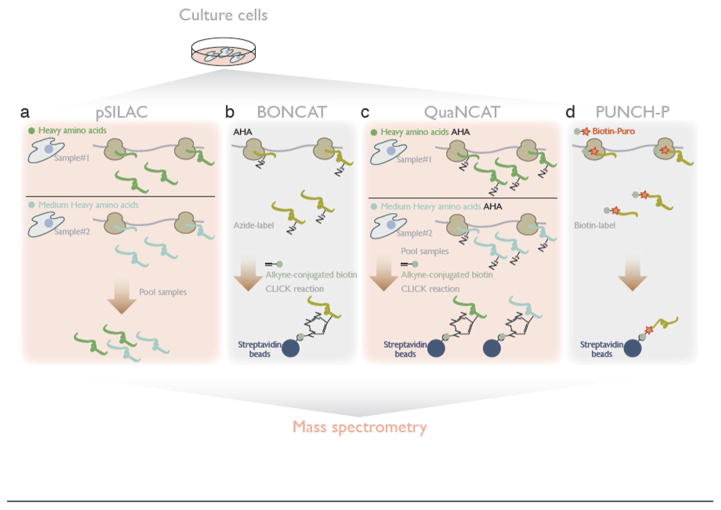
Figure 2. Mass spectrometry-based methods for measuring differential translation changes across proteome.
a. Pooling proteins from two samples, in the pSILAC (pulsed stable isotope labeling by amino acid in cell culture) method, provides quantitative mass spectrometry of protein produced during the labeling period.
b. In BONCAT (bio-orthogonal non-canonical amino acid tagging), CLICK-reactive bio-orthogonal amino acids [e.g. AHA (azidohomoalanine)] are incorporated into protein and subsequent biotin-tagging allows affinity purification of newly synthesized protein, for wider coverage of the proteome.
c. QuaNCAT (quantitative noncanonical amino acid tagging) combines pSILAC and BONCAT for quantitative mass spectrometry with broader coverage.
d. PUNCH-P captures polypeptides from active translation through biotin-conjugated puromycin and subsequent purification by streptavidin beads.