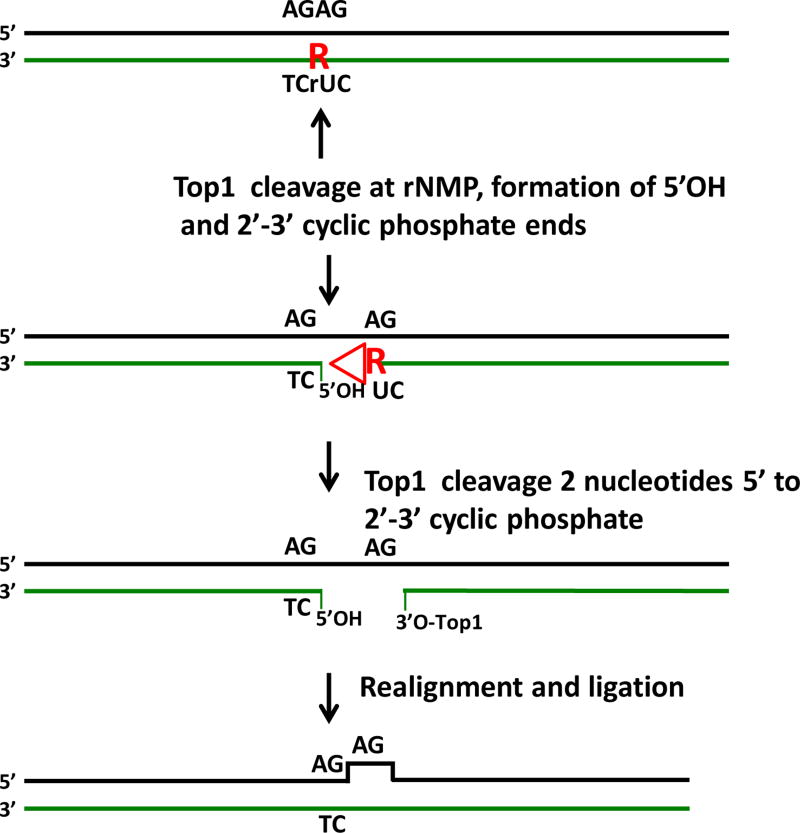
Figure 2.
Top1 processing of rNMP residues. An embedded rNMP residue (red R) is shown in a repeated sequence AGAG:TCrUC. In the absence of the RER process Top1 can cleave at rNMP residues in duplex DNA. Top1 cleaves 3’ to the residue, and in completeting the Top1 cleavage cycle, generates a 5’OH end and a 2’-3’ cyclic phosphate end (red triangle) through nucleophilic attack by the 2’OH group of the ribose sugar. A second cleavage by Top1 2 nucleotides from the 2’–3’ cyclic phosphate end releases this blocked end and the ribose residue. If the resulting gap is in a repeated sequence as shown, slippage alignment can occur during completion of the Top1 reaction and the gap is resealed by Top1, leading to a −2 deletion on the strand that originally contained the rNMP residue.