Figure 1.
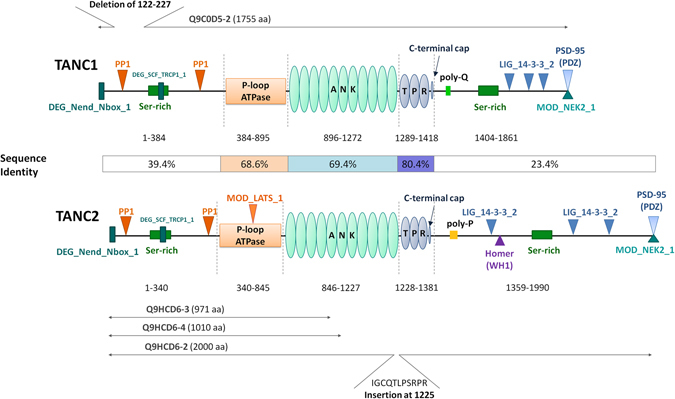
Sequence analysis of TANC proteins. An overview of TANC family domain architecture is here reported. Both TANC proteins are characterized by a putative P-loop NTPase domain (orange), an Ankyrin repeat containing domain (light teal) and a tetratricopeptide repeat region (blue). For each domain, the sequence boundaries and sequence identity between the two proteins are indicated. Conserved linear motifs are represented as follow: PDZ binding sequences (light blue triangles); PP1 docking motif (RVxF) (orange triangles); degrons (DEG_Nend_Nbox_1 and DEG_SCF_TRCP1_1) (deep teal rectangles); 14_3_3 binding sites (LIG_14-3-3_2) (blue triangles); Homer binding motif (LIG_EVH1_1) (Purple triangle); LATS1 kinase (light orange triangle); NEK2 phosphorylation motif (MOD_NEK2_1) (Teal triangle). Serine-rich regions are represented with green rectangles (TANC1 residues 170-243 and 1659–1689; TANC2 residues 125–189 and 1775–1865). The TANC1 glutamine-rich region (poly-Q) region and TANC2 glutamine/proline rich region (polyP) are in light green and yellow respectively. Alternative TANC protein isoforms are reported in grey. The TANC1 isoform Q9C0D5-2 (1755 residues) is missing the region 122-227. The TANC2 isoform Q9HCD6-2 is longer (2000 residues) due to an insertion at position 1225 (I > IGCQTLPSRPR). Q9HCD6-3 (971 residues) is truncated at residue 97 with different substitution in the region from position 944 to 971 (VDHLDKNGQCALVHAALRGHLEVVKFLI > VLAAQLCCFSSLFLYFRCILFLISSVTS). Q9HCD6-4 (1,010 residues) is truncated at residue 1011 with different substitution in the region from position 1006 to 1010 (IVSYL > VRSRQ).
