Figure 2.
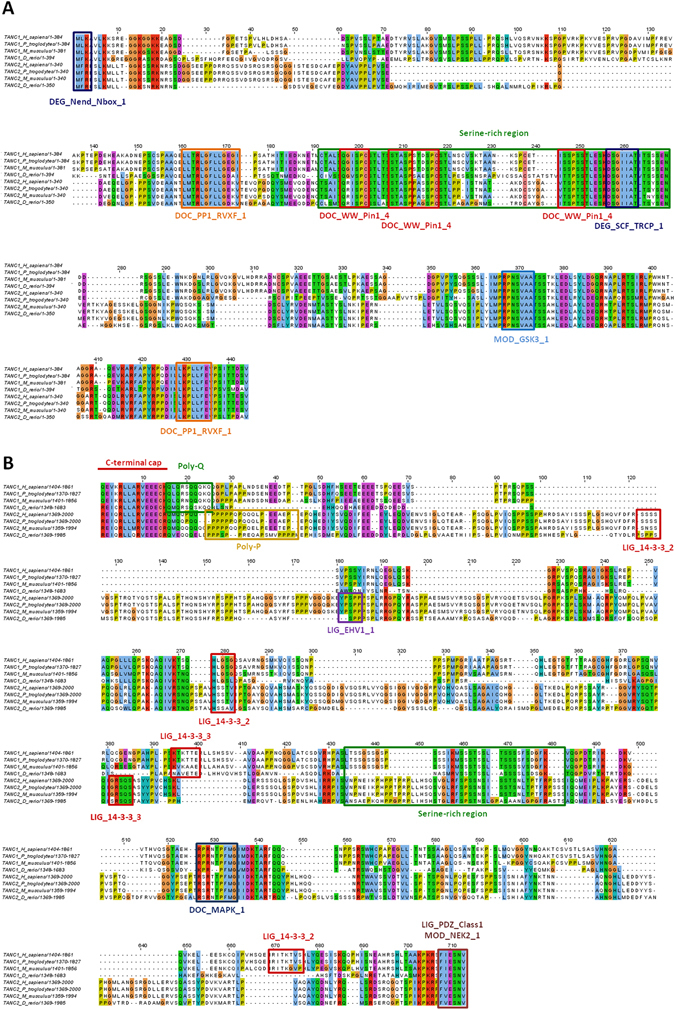
Multiple alignments of TANC N- and C- termini. Colour code based on Clustalx scheme. Linear motifs identified by ELM analysis are reported: DEG_Nend_Nbox_1: N-terminal motif that initiates protein degradation by binding to the N-box of N-recognins; DOC_PP1_RVXF_1: Protein phosphatase 1 catalytic subunit (PP1c) interacting motif; DOC_WW_Pin1_4: IV WW domain interaction motif; MOD_GSK3_1: GSK3 phosphorylation recognition site; MOD_NEK2_1: NEK2 phosphorylation motif; DEG_SCF_TRCP1_1: DSGxxS phospho-dependent degron recognized by F box protein of the SCF-betaTrCP1 complex; LIG_14-3-3_2: 14-3-3-binding motif; DOC_MAPK_1: MAPK docking motifs; LIG_PDZ_Class_1: PDZ-binding motif; LIG_14-3-3_3: 14-3-3-binding motif; LIG_EVH1_1: Proline-rich motif binding to signal transduction class I EVH1 domains. (A). N-terminus, (B). C-terminus.
