Figure 10.
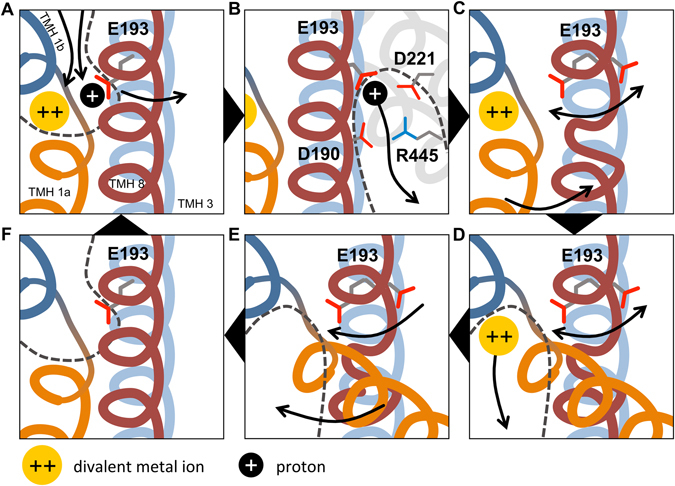
Proposed mechanistic model of the transport cycle of hDMT1. Transmembrane helices are shown color-coded: TMH 1a (orange), TMH 1b (dark blue), TMH 3 (light blue), TMH 8 (red), with TMH 3 located behind TMH 8. Divalent metal ion substrate is shown as yellow disc; proton as black disc. Water-accessible vestibules are indicated with dashed lines. Arrows indicate movements of components. (A) Substrate binding happens in the high-pK a state of E193, likely in an ordered manner. After substrates bind, a conformational switch is proposed to occur, bringing the protein into an inward-open-like occluded state, where E193 flips over to a low-pK a conformation. (B) In the low-pK a conformation, proton dissociation occurs possibly along a pathway flanked by D221, D190 and R445 (D153, D124 and R360 in ScaDMT, respectively). (C) The loss of the proton induces three key conformational changes: the partial unwinding of TMH 8 near residues 410–417 (corresponding to residues 325–330 in ScaDMT), increased flexibility of the E193 side-chain and the opening of the intracellular gate. At this point, substrate-dependent uncoupled proton leak could happen at high proton concentrations by jumping back to state A, if transition to state D is slow enough. (D) After gate opening, the metal ion substrate is solvated and leaves the binding site. (E) Local helix unwinding in TMH 8 facilitates the recovery of E193 into the high-pK a state, and closure of the intracellular gate happens. (F) The high-affinity outward-open state is recovered. Additionally, this mechanism also allows for the uniport of metal ions in the absence of protons through cycle A-D-E-F-A.
