Abstract
Genetically engineered (GE) rice endogenous epsps (5-enolpyruvoylshikimate-3-phosphate synthase) gene overexpressing EPSPS can increase glyphosate herbicide-resistance of cultivated rice. This type of epsps transgene can enhance the fecundity of rice crop-weed hybrid offspring in the absence of glyphosate, stimulating great concerns over undesired environmental impacts of transgene flow to populations of wild relatives. Here, we report the substantial alteration of phenology and fitness traits in F1-F3 crop-wild hybrid descendants derived from crosses between an epsps GE rice line and two endangered wild rice (Oryza rufipogon) populations, based on the common-garden field experiments. Under the glyphosate-free condition, transgenic hybrid lineages showed significantly earlier tillering and flowering, as well as increased fecundity and overwintering survival/regeneration abilities. In addition, a negative correlation was observed between the contents of endogenous EPSPS of wild, weedy, and cultivated rice parents and fitness differences caused by the incorporation of the epsps transgene. Namely, a lower level of endogenous EPSPS in the transgene-recipient populations displayed a more pronounced enhancement in fitness. The altered phenology and enhanced fitness of crop-wild hybrid offspring by the epsps transgene may cause unwanted environmental consequences when this type of glyphosate-resistance transgene introgressed into wild rice populations through gene flow.
Introduction
The unpredicted potential environmental impact caused by transgene flow from genetically engineered (GE) crops to their cross-compatible wild relatives has stimulated tremendous debates and studies over the last decades1–3. Wild relative populations that have acquired a strongly fitted transgene through gene flow likely changes their evolutionary potential, resulting in unwanted environmental/ecological consequences4–7. It is therefore essential to properly assess the environmental impact of transgene flow before the commercialization of any GE crops. One of the key points to assess such environmental impact is to determine fitness of a transgene introgressed into wild populations7, 8, provided that the frequency of crop-to-wild gene flow is known. Many studies have been carried out to determine the fitness effect of a transgene under the controlled field-experimental conditions, involving crop-wild hybrid lineages. These studies included hybrid descendants derived from crosses between squash-wild gourd9, 10, maize-teosinte11, cultivated-wild sunflowers12, and cultivated-wild/weedy rice13–18. The generated data provided useful information for the biosafety assessment of environmental impact caused by transgene flow.
Herbicide-resistant GE crops are predominantly cultivated in the world because of their evident advantages to reduce the crop production costs through effective agriculture weed control19, 20. To date, GE crops containing an herbicide-resistance transgene(s) have occupied nearly 90% of the global arable lands where all GE crops with different transgenes are grown, including GE soybean, cotton, maize, and canola21. Breeders are enthusiastic to develop herbicide-resistant rice (Oryza sativa L.)—a very important world cereal crops, to meet the challenge of world food security by resolving the great problems of weed control, particularly under the situation of shifting rice cultivation from traditional transplantation to direct seeding22, 23. In direct-seeding fields, rice production faces more serious problems for weed control, particularly for weedy rice (O. sativa f. spontanea, referred to as WDR hereafter) that belongs to the same biological species of cultivated rice24, 25. Supposedly, the application of herbicide-resistant rice can effectively reduce the infestation of weedy rice23, 26, although this proposal faces challenges16, 20, 23. That is, the introgression of herbicide-resistance transgenes from GE rice into WDR populations through spontaneous gene flow and its social/environmental impact by increased weed problems becomes a great biosafety concern3. For WDR that mostly infests rice fields, the rapid spread of an introgressed herbicide-resistance transgene to its populations is expected because of the strong selective pressure from herbicide application in rice fields. WDR populations with substantially increased herbicide-resistance and weediness have been found in Clearfield® rice fields, causing tremendous weed problems27, 28. The concerns over the spread of herbicide-resistance transgenes to WDR populations is reasonable in the development and commercialization of herbicide-resistant GE rice.
However, it is argued that the spread of herbicide-resistance transgenes to wild rice (Oryza species) populations may not cause similar problems as in WDR populations, because wild rice populations occur in nature habitat where no herbicides are used and therefore selection pressure from herbicides does not exist29, 30. In contrast, herbicide-resistance transgenes are considered to be neutral or even have costs in plants under the herbicide-free environmental conditions31, 32. Therefore, wild rice populations that have acquired an herbicide-resistance transgene through gene flow may not have any benefit due to the lack of selective pressure from herbicides33. Nevertheless, a recent study reported significantly increased fecundity and ratios of tryptophan concentration and photosynthesis in crop-weed hybrid lineages containing an herbicide-resistance transgene, at the absence of herbicide16. This transgene is an engineered rice endogenous epsps (5-enolpyruvoylshikimate-3-phosphate synthase) gene that overexpresses EPSPS. The GE rice was originally developed to confer tolerance to the glyphosate herbicide34, but unexpectedly the epsps transgene provided substantial benefits to WDR for plant growth and seed production16. This phenomenon poses a question about the transgene that overproduces EPSPS. Will the epsps herbicide-resistance transgene introgressed into any of the wild rice species produce the same fitness effect to their populations?
The perennial common wild rice (O. rufipogon Griff., referred to as WR hereafter) is the direct ancestor of Asian cultivated rice and one of the wild species in the genus Oryza (Poaceae). WR is distributed in the tropics and subtropics of monsoon Asia, with its northernmost border in Jiangxi province of China35. WR can reproduce sexually through seeds, or asexually through propagule or ratooning36, 37. It is widely recognized that WR is important germplasm for the genetic improvement of cultivated rice. For example, the well-known hybrid rice breeding program was benefited from the discovery and use of a male sterility (ms) gene from WR38, 39, demonstrating the importance of germplasm in WR gene pool40. However, WR is under threats due to the rapid growth of human population and urbanization, dramatic change in agriculture land uses, and intensive human disturbances41, 42. Also, massive and continued introgression of cultivated rice genes and transgenes into WR have posed a great challenge on the existence of WR populations3, 43. Altogether, identified gene flow from cultivated rice to WR populations in the controlled experiments44, 45 and population genetic studies46 indicated the high probability of transgene introgression from GE rice to WR. In addition, the diverse genetic variability among WR populations may result in different fitness responses of WR recipients to the same transgene, as revealed in a recent study of crop-weed hybrids containing insect-resistance transgenes18.
We produced F1-F3 crop-wild isogenic hybrid lineages with or without the epsps transgene, derived from artificial crosses between an epsps GE rice line and two WR populations. The objectives of this study were to address the following questions. (1) Does the over-expressing epsps transgene change the life-cycle traits of crop-wild hybrid descendants in the glyphosate-free environment? (2) Does the epsps transgene increase the fecundity and over-winter survival of crop-wild hybrid descendants? (3) Does the genetic background of transgene recipients affect the fitness of the epsps transgene in different types of rice parents, including WR, WDR, and cultivated rice? The answer of these questions will help us to appropriately estimate the potential environmental/ecological consequences caused by introgression of the overexpressing epsps herbicide-resistance transgene into WR populations, likely also to predict the potential consequences for other crop-wild transgene introgressions.
Results
More tillers and earlier flowering with increased seed sets in transgenic hybrid lineages
F1 and F2 hybrid lineages with the epsps transgene showed a greater number of tillers and earlier flowering time than their isogenic controls without the transgene (Fig. 1a, b, c and d; Tables S1–S3). Consequently, ratios of seed sets in the transgenic hybrid lineages showed significant increase, compared to their non-transgenic counterparts (Fig. 1e and f; Table S1). An obvious negative correlation was observed between ratio of seed set and days from seed germination to flowering in F2 hybrid descendants (Fig. S1a and b). In addition, no differences were observed for plant height between transgenic and non-transgenic hybrid lineages (Table 1).
Figure 1.
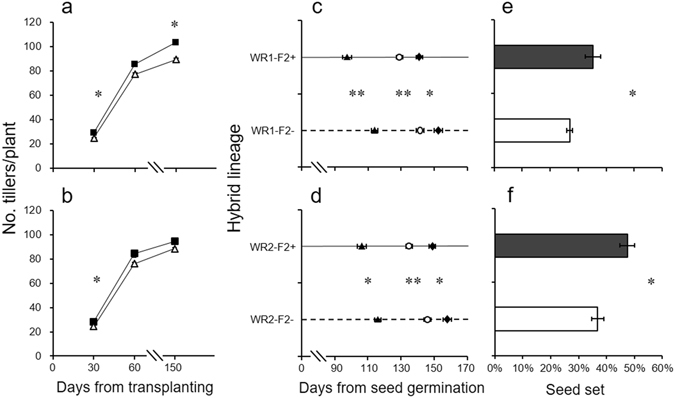
Number of tillers per plant (a: WR1-F2; b: WR2-F2); flowering time (c: WR1-F2; d: WR2-F2); and seed set ratios (e: WR1-F2; f: WR2-F2) of F2 transgenic (solid squares, dark grey columns) and non-transgenic (empty triangles, white columns) hybrid lineages in pure planting. Solid triangles, empty circles, and solid diamonds (in c and d) indicate days at which 1%, 30%, and 50% plants being flowered, respectively. The comparisons were made between transgenic and non-transgenic F2 hybrid lineages in pure-planting based on independent t-test (N = 6). Bars represent standard error. * or ** indicates significances at the levels of P < 0.05 or P < 0.01, respectively.
Table 1.
Two-way ANOVAs for the effects of transgene (transgenic vs. non-transgenic), wild parent (WR1 vs. WR2), and their interactions on life-cycle fitness related traits of F1-F3 rice crop-wild hybrid descendants.
| Trait | Transgene (T) | Wild parent (WR) | T × WR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| df | F | P | df | F | P | df | F | P | |
| F1 experiment | |||||||||
| Plant height | 1 | 0.076 | 0.786 | 1 | 0.011 | 0.916 | 1 | 0.158 | 0.695 |
| No. tillers per plant-30 days | 1 | 22.293 | 0.000 | 1 | 12.107 | 0.002 | 1 | 0.957 | 0.340 |
| No. tillers per plant-60 days | 1 | 22.212 | 0.000 | 1 | 0.053 | 0.820 | 1 | 0.385 | 0.542 |
| No. tillers per plant-150 days | 1 | 26.674 | 0.000 | 1 | 0.064 | 0.803 | 1 | 0.463 | 0.504 |
| Days for 1% plants to flower | 1 | 7.347 | 0.013 | 1 | 1.837 | 0.190 | 1 | 0.000 | 1.000 |
| Days for 30% plants to flower | 1 | 6.792 | 0.017 | 1 | 6.792 | 0.017 | 1 | 0.189 | 0.669 |
| Days for 50% plants to flower | 1 | 5.902 | 0.025 | 1 | 13.279 | 0.002 | 1 | 0.000 | 1.000 |
| No. panicles per plant | 1 | 9.205 | 0.007 | 1 | 0.720 | 0.406 | 1 | 0.208 | 0.654 |
| No. seeds per plant | 1 | 3.541 | 0.074 | 1 | 12.544 | 0.002 | 1 | 0.367 | 0.551 |
| 1000-seed weight | 1 | 0.779 | 0.388 | 1 | 0.819 | 0.376 | 1 | 0.667 | 0.424 |
| Ratio of seed set | 1 | 5.760 | 0.026 | 1 | 33.180 | 0.000 | 1 | 3.687 | 0.069 |
| Ratio of tiller regeneration | 1 | 14.861 | 0.000 | 1 | 0.763 | 0.384 | 1 | 2.213 | 0.140 |
| F2 experiment | |||||||||
| Plant height | 1 | 0.592 | 0.451 | 1 | 5.910 | 0.025 | 1 | 0.407 | 0.531 |
| No. tillers per plant-30 days | 1 | 12.593 | 0.002 | 1 | 0.161 | 0.693 | 1 | 0.040 | 0.844 |
| No. tillers per plant-60 days | 1 | 3.278 | 0.085 | 1 | 0.055 | 0.816 | 1 | 0.001 | 0.976 |
| No. tillers per plant-150 days | 1 | 9.428 | 0.006 | 1 | 2.077 | 0.165 | 1 | 1.498 | 0.235 |
| Days for 1% plants to flower | 1 | 23.719 | 0.000 | 1 | 4.246 | 0.053 | 1 | 1.719 | 0.205 |
| Days for 30% plants to flower | 1 | 25.508 | 0.000 | 1 | 5.092 | 0.035 | 1 | 0.104 | 0.751 |
| Days for 50% plants to flower | 1 | 17.474 | 0.000 | 1 | 7.578 | 0.012 | 1 | 0.260 | 0.616 |
| No. panicles per plant | 1 | 7.822 | 0.011 | 1 | 0.018 | 0.894 | 1 | 0.202 | 0.658 |
| No. seeds per plant | 1 | 9.171 | 0.007 | 1 | 46.538 | 0.000 | 1 | 0.067 | 0.799 |
| 1000-seed weight | 1 | 0.021 | 0.887 | 1 | 5.953 | 0.024 | 1 | 0.187 | 0.670 |
| Ratio of seed set | 1 | 17.477 | 0.000 | 1 | 24.342 | 0.000 | 1 | 0.240 | 0.630 |
| Ratio of tiller regeneration | 1 | 6.157 | 0.015 | 1 | 3.375 | 0.069 | 1 | 4.255 | 0.042 |
| F3 experiment | |||||||||
| Seed germination ratio-0 day | 1 | 1.023 | 0.341 | 1 | 2.527 | 0.151 | 1 | 0.002 | 0.963 |
| Seed germination ratio-20 days | 1 | 70.621 | 0.000 | 1 | 79.724 | 0.000 | 1 | 1.103 | 0.324 |
| Seed germination ratio-40 days | 1 | 65.154 | 0.000 | 1 | 13.462 | 0.006 | 1 | 10.560 | 0.012 |
| Seed germination ratio-60 days | 1 | 21.437 | 0.002 | 1 | 35.438 | 0.000 | 1 | 6.509 | 0.034 |
Df, degree of freedom; F, F values; P, level of significance.
Two-way ANOVAs showed significant effects of a transgene (T) and wild parents (WR) on the number of tillers per plant (at different growth stages), days to flowering, and ratios of seed sets in both F1 and F2 hybrid descendants. However, no significant interaction effect was detected (Table 1). In the pure planting mode, significant increases were detected for the number of tillers per plant in F1 transgenic hybrids throughout the growth stages (with 19~38% increase); but significant increases were mainly detected at the early growth stages in F2 transgenic hybrid lineages (with 16~17% increase) (Fig. 1a and b; Table S1). In the mix-planting mode with 30 × 30 cm spacing between plants, significant differences were also detected for the number of tillers per plant, but with more prominent differences between transgenic and non-transgenic hybrid lineages (with 26~49% increase in F1 and 9~24% increase in F2) (Fig. 2a and b; Tables S2 and S3). Noticeably, the increase in number of tillers was more pronounced in the 30 × 30 cm mix-planting plot than that of other plots with lower densities (Fig. 2a and b; Tables S2 and S3). In addition, transgenic hybrid plants flowered significantly earlier (3 days for F1 and 9~15 days for F2) than their non-transgenic counterparts (Fig. 1c and d; Table S1). Consequently, transgenic hybrid plants had significantly higher ratios of seed sets (29% in F1 derived from WR1 and 29–30% in F2) than their non-transgenic counterparts in pure planting (Fig. 1e and f; Table S1). The ratios of seed sets were negatively correlated with the days to flowering in F2 hybrid lineages with or without the transgene (Fig. S1a and b), suggesting the possible contribution of earlier flowering to greater seed sets.
Figure 2.
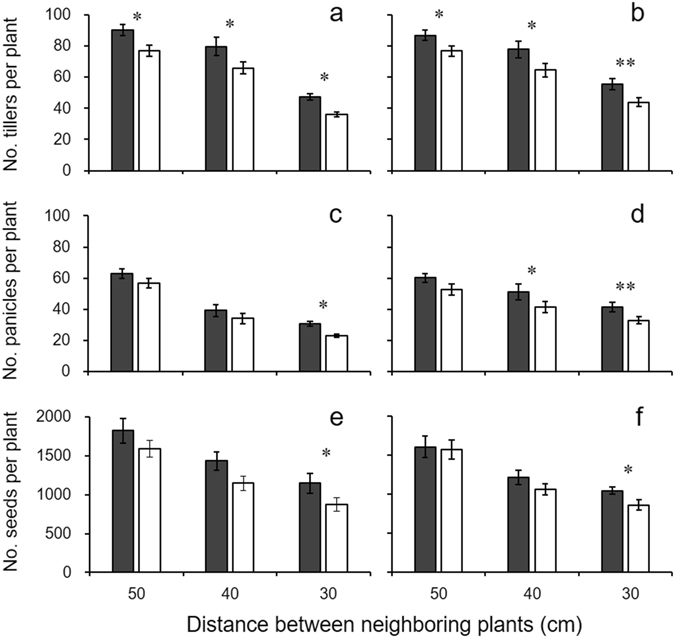
Number of tillers (a, b), panicles (c, d), and filled seeds (e, f) per plant in F1 transgenic (dark grey columns) and non-transgenic (white columns) crop-wild hybrids derived from WR1 (left panel) and WR2 (right panel), in mixed-planting plots (30 cm, 40 cm, 50 cm plant spacing) with different densities. The comparisons were made between transgenic and non-transgenic F1 hybrids in mix planting based on paired t-test (N = 6). Bars represent standard error. * or ** indicates significances at the levels of P < 0.05 or P < 0.01, respectively.
Increased fecundity in transgenic hybrid lineages
Both F1 and F2 transgenic hybrid lineages showed significantly increased fecundity as indicated by the number of panicles and seeds per plant compared to their non-transgenic counterparts in mix-planting plots with 30 × 30 cm spacing (Figs 2c, d, e, f and 3b and d; Tables S2 and S3). The hybrid lineage with WR1 as the wild parent also showed significantly increased fecundity in pure planting (Fig. 3a and c; Table S1). In addition, no differences were detected for 1000-seed weight between transgenic and non-transgenic hybrid lineages (Table 1). Two-way ANOVAs showed a significant effect of transgene (T) on the number of panicles per plant in F1 and F2 crop-wild hybrid descendants (Table 1). Wild parent (WR) had significant effect on the number of seeds per plant in F1-F2 and 1000-seed weight in F2 hybrid descendants (Table 1). No significant interaction effect was detected (Table 1). In the pure planting mode, 17~22% (F1) and ~13% (F2) increases were detected in number of panicles per plant in transgenic hybrid lineages (Fig. 3a; Table S1); meanwhile, ~27% increase was detected for the number of filled seeds per plant in F2 transgenic hybrid lineages (Fig. 3c; Table S1). In the mix-planting mode with different densities, significant increase in number of panicles (with 10~33% increase) and filled seeds (with 21~38% increase) per plant was mainly observed in the 30 × 30 cm plots in F1 and F2 transgenic hybrid lineages (Figs 2c, d, e, f and 3b and d; Tables S2 and S3). It seems that with the increase in cultivation densities, the extent of increases in panicles and filled seeds per plant between transgenic and non-transgenic plants became more substantial, suggesting the competitive effect for fecundity (Fig. 2c, d, e, and f).
Figure 3.
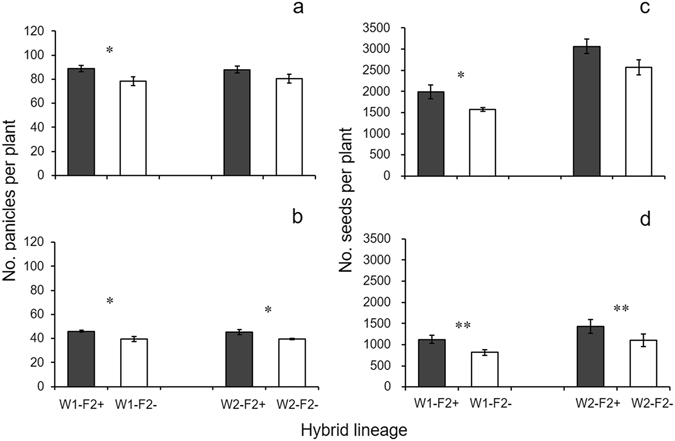
Number of panicles (a, b) and filled seeds (c, d) per plant of F2 transgenic (dark grey columns) and non-transgenic (white columns) crop-wild hybrid lineages in pure-planting (a, c) and mix-planting plots (b, d). The comparisons were made between transgenic and non-transgenic F2 hybrid lineages in mix planting (30 cm) based on paired t-test (N = 6). Bars represent standard error. * or ** indicates significances at the levels of P < 0.05 or P < 0.01, respectively.
In addition, the two transgenic events (EP3 and EP4) showed similar extent of glyphosate resistance and increased number of panicles and filled seeds per plant (Table S4), suggesting the observed differences between transgenic and non-transgenic lineages were not the result of transgene insertion effect.
Enhanced buried-seed germination and over-winter regeneration in transgenic hybrid lineages
The transgenic hybrid lineages had higher germination ratios for soil-buried seeds (F3) and tiller regeneration ratios overwinter (F1 and F2) than non-transgenic counterparts, especially for those derived from WR1 (Fig. 4a, b, c and d; Tables 1 and S1).
Figure 4.

Germination ratios of buried-seeds (a: WR1-F3, b: WR2-F3) and ratios of tiller regeneration (c: WR1-F1 and WR1-F2, d: WR2-F1 and WR2-F2) in transgenic (dark grey columns) and non-transgenic (white columns) crop-wild hybrid lineages. The comparisons were made between transgenic and non-transgenic hybrid lineages based on independent t-test (N = 3 for seed germination ratio; N = 6 for tiller regeneration ratio). Bars represent standard error. * or ** indicates significances at the levels of P < 0.05 or P < 0.01, respectively.
Two-way ANOVAs showed significant effect of transgene (T) on the ratios of seed germination (after being buried for 20, 40, and 60 days) and tiller regeneration. Wild parent (WR) had significant effect on ratios of buried-seed germination. Significant effect was detected for interaction between T and WR on the ratios of buried-seed germination (only for 40 and 60 days) and tiller regeneration (Table 1). No significant differences in germination ratios were detected between transgenic and non-transgenic hybrid lineages before seed burial. However, transgenic hybrid lineages (from WR1) showed 26%, 45%, and 38% higher seed germination ratio than their non-transgenic counterparts, 20, 40, and 60 after days after burial (Fig. 4a). A similar trend was also observed in hybrid descendants derived from WR2 (Fig. 4b). Transgenic F1 and F2 hybrid lineages derived from WR1 showed 55% and 275% higher tiller regeneration ratios compared to their non-transgenic counterparts (Fig. 4c). However, no significant differences were detected between hybrid lineages derived from WR2 (Fig. 4d), indicating apparent maternal influences.
Differences in fitness-related traits affected by endogenous EPSPS level of transgene recipients
The content of endogenous EPSPS was varied significantly among different transgene recipients (WR, WDR, and cultivated rice parents) at different growth stages (Fig. 5). The degree of fitness differences (as indicated by the ratios of increased panicles and seeds per plant) between transgenic and non-transgenic hybrid lineages varied substantially among different transgene recipients. A weak negative correlation was observed between the fitness differences caused by the incorporation of the epsps transgene and the content of endogenous EPSPS of different types of the transgene recipients (Fig. 5).
Figure 5.

Correlation between the ratios of increased panicles (a, b, and c) or seeds (d, e, and f) and the content of endogenous EPSPS proteins in different transgene recipients (parents) at 60 (a and d), 100 (b and e), and 160 (c and f) days after seed germination. Solid diamonds: wild rice (WR1); empty diamonds: wild rice (WR2); empty circles: weedy rice (WRD1); solid circles: weedy rice (WRD2), and solid triangles: cultivated rice (Minghui-86).
The two WDR populations expressed a low level of endogenous EPSPS (0.05~0.33%) at different growth stages. Accordingly, their transgenic lineages showed a more substantial increase in the number of panicles (17~34%) and filled seeds (55~57%) per plant (Fig. 5). In contrast, the two WR populations expressed a relatively high level of endogenous EPSPS (0.23~0.72%), and their transgenic counterparts showed a less increase in the number of panicles (9~13%) and filled seeds (19~27%) (Fig. 5). The parental rice line (Minghui-86) showed a moderate level of endogenous EPSPS and fecundity change (Fig. 5) compared to the WR and WDR populations. These results suggested the possible association, although not strong, between the levels of endogenous EPSPS expression and differences in fitness caused by the epsps transgene.
Discussion
We found evidently altered phenological characteristics (e.g., higher tillering rates and earlier flowering), increased fecundity (more seeds and higher seed-set ratios), and enhanced ability of overwinter survival for stocks/tillers in the crop-wild rice hybrid lineages containing an epsps glyphosate-resistance transgene, based on our three-year common-garden experiments. These results suggest that the transgene over-expressing epsps can change the life-cycle characteristics and increase fitness of crop-wild rice hybrid descendants in the glyphosate-free environment, similar with that reported by Wang et al.16 in a study with the same epsps transgene in crop-weed hybrid descendants. In addition, we also detected differences in the base-line expression level of the EPSPS protein encoded by the endogenous epsps gene in transgenic recipients (parents), which influenced the fitness effect of the epsps transgene in their corresponding crop-wild/weed hybrid lineages. This result suggests that the genetic background of transgene recipient populations can considerably affect fitness of a transgene, which is similar with that in a study involving two insect-resistance transgenes (Bt, Bt/CpTI)18. Altogether, these findings answered our questions raised up earlier that crop-to-wild introgression of a glyphosate-resistance transgene overexpressing epsps through gene flow will significantly change the life-cycle fitness of WR populations in the herbicide-free habitats. In general, such altered life-cycle fitness of crop-wild hybrid descendants caused by the transfer of a transgene overexpressing epsps to wild relatives will likely generate unwanted environmental impact, including the creation of more invasive agricultural weeds and losses of genetic integrity of wild relative species of crops.
The higher tillering rates at different growth stages and earlier flowering time will likely increase the competitiveness of the hybrid plants that acquired the epsps transgene and shift their reproductive period some days ahead. The increased tillering rate detected in crop-wild hybrid lineages from this study is similar as that reported in the previous study based on analyses of crop-weed hybrid lineages16. Our findings suggest that transgenes overexpressing EPSPS may promote the persistence and spread of crop-wild and crop-weed transgenic hybrid descendants in rice in the glyphosate-free environment. However, some studies did not show fitness changes in mutational herbicide-resistant weeds with multiple copies of endogenous epsps genes47–49, suggesting the complex of the epsps genes in terms of the fitness change in plants. Further studies are required to reveal the underlying mechanisms. It is well known that EPSPS encoded by the epsps gene is a key enzyme in the shikimic acid pathway that is closely associated with plant growth through producing necessary aromatic amino acids, lignin, flavonoids, phenolics, and other secondary metabolic substances50, 51. The overexpressed EPSPS by transgene will not only improve the glyphosate-resistance of the transgene recipient52, but also affect their phenotype associated with the change in shikimic acid pathway. Probably, the observed higher tillering rates and earlier flowering of transgenic crop-wild hybrid lineages in our study was caused by the exceeded production of EPSPS that promoted the biosynthesis of metabolic substances in the downstream pathways53, 54. Consequently, the altered phenological characteristics advanced the reproductive period of transgenic hybrid plants, which may provide more opportunities for hybrid lineages to reproduce sexually under more favorable climate conditions (e.g., temperature and moistures) and to circumvent cold stress before winter comes. These changes resulted in the increases in seed production and seed-set ratios, as observed in our experiments. In addition, more vigorous plant growth as indicated by increased tillers can enhance the competitive ability of epsps transgenic hybrid plants, compared to those without the transgene. This hypothesis gains a support from the mix-planting plots with dense cultivation of plants in our experiments, where differences in the number of tillers, panicles, and filled seeds between transgenic and non-transgenic plants were more outstanding. The increased competitiveness and altered reproductive period of transgenic hybrid plants may eventually affect the life-cycle fitness of the WR populations.
The fecundity or seed production of hybrid lineages as indicated by the increased number of panicles and well-developed seeds per plant was considerably affected by the altered phenological characteristics as indicated above. Obviously, the greater number of panicles per plant observed in transgenic hybrid lineages is associated with their greater number of tillers per plants and enhanced competitiveness at the early growth stages. The increased number of seeds per plant found in transgenic crop-wild hybrid lineages is more likely attribute to both greater number of panicles per plant and the increased seed-set ratios, as affected by the advanced reproduction in slightly warmer conditions. The increased fecundity brought by the epsps transgene was also observed in other studies. For example, the epsps transgene increased fecundity of F1-F2 crop-weed hybrid lineages in rice16. The yield increase was also recorded in the GE glyphosate-resistant (CP4) soybean cultivars, compared with their non-GE counterparts in the absence of glyphosate55, although the authors attributed the observed yield increase to the improved genetic background for the GE cultivars. In addition, a study of GE glyphosate-resistant wheat lines containing an epsps (CP4) transgene driven by rice actin1 and 35S promoters showed considerable increases in the grain yield without glyphosate applications in an experiment56. All these studies demonstrate that the epsps transgene can increase fecundity of GE crops and particularly crop-wild/weed hybrid descendants in the environment without the application of glyphosate herbicides, which will promote the persistence and spread of the transgenes.
The significantly enhanced overwinter ability of transgenic hybrid descendants (particularly those derived from WR1), as measured by seed germination after being buried in the soil and tiller survival/regeneration through winters, indicates enhanced tolerances of hybrid plants to environmental stresses by introducing the epsps transgene. The increased germination ratios of transgenic seeds after being buried in soils is probably due to their increased tolerance to the biotic and abiotic stress in the soil, although the underlying mechanisms need further studies to be revealed. In addition, the enhanced ability of tiller survival/regeneration in transgenic hybrid lineages suggests their enhanced tolerance to cold stress in winter, promoting the initial competitive ability of transgenic hybrid seedlings to occupy more territories. WR populations can propagate vegetatively mainly through tillers/stocks and ratoons during the winter, particularly for the large populations occurring in deep-water habitats57. The overwintering survival/regeneration ability of the transgenic plants is critical for the successful maintenance and expanding of O. rufipogon populations in the next year. It is apparent that the enhanced ability of vegetative propagation through winter, together with the increased fecundity of crop-wild hybrid descendants containing the epsps transgene, will considerably increase the life-cycle fitness advantages, resulting in unwanted environmental consequences.
In addition, our results of significant differences in base-line endogenous EPSPS contents among different WR, WDR populations and cultivated rice line indicate the influences of genetic background of transgene recipients on the fitness effect. For example, the two WDR populations with a lower level of endogenous EPSPS showed much more pronounced enhancement in fitness (e.g., number of panicle and seeds per plant) in crop-weed hybrid descendants than two WR populations with a higher EPSPS level. Probably, WDR plants with relatively lower level of endogenous EPSPS response more sensitively to the epsps transgene, leading to more dramatic changes in fitness in transgenic crop-weed hybrid descendants, and vice versa. A similar phenomenon was also found in another study, in which different fitness effects of insect-resistance transgenes were detected in crop-weed hybrid descendants derived from crosses between a GE rice line and five WDR populations with different origins, respectively18. Therefore, differences in the genetic background should be considered in the biosafety assessment of transgene introgression into different wild/weedy recipient populations, following the case-by-case principle2, 3, 5.
In conclusion, our study demonstrated the altered phenology and enhanced life-cycle fitness of crop-wild hybrid descendants containing the epsps transgene in the glyphosate-free environment. The fitness change may cause unwanted environmental consequences, once this type of herbicide-resistance transgene introgressed into WR populations through gene flow. On one hand, WR individuals/populations that have picked up the epsps transgene may become more aggressive weeds because of their increased fitness benefit in natural habitats, although the problem may become more serious in the agricultural ecosystems where glyphosate herbicides are applied1, 3, 6, 7, 23. On the other hand, introgression of the epsps transgene may also affect the genetic integrity of local WR populations and cause their reduction or even extinction due to genetic swamping and selective sweep effects1, 3, 43, 58. Therefore, it is necessary to design proper strategies to effectively assess and manage the potential risks caused by introgression of transgenes that can increase weediness and invasiveness of wild relative populations.
Materials and Methods
Production of crop-wild hybrid lineages
An herbicide-resistant transgenic rice (Oryza sativa L.) line (EP3), its non-transgenic rice parent (Minghui-86), and two WR populations (WR1, WR2) were used to generate F1-F3 crop-wild hybrid descendants. The EP3 transgenic line containing a GE endogenous epsps gene from rice was produced via the Agrobacterium-mediated transformation from Minghui-8616, 59. The EP3 line used for hybridization was a T5 generation homozygous for the epsps transgene that was resistant to glyphosate34. The non-transgenic Minghui-86 is a widely-used rice variety in China. The two O. rufipogon populations were collected from Dongxiang in Jiangxi province (WR1) and Suixi in Guangdong province (WR2), China. To avoid the possible gene insertion effects at different loci on phenotypes, we analyzed glyphosate resistance and two fecundity-related traits (number of panicles and filled seeds per plant) of two independent transgenic rice lines (EP3 and EP4), using their non-transgenic parent (Minghui-86) as a control.
For creating crop-wild hybrid lineages, we produced F1 transgenic and non-transgenic hybrids by hand pollination of WR plants (more than 20 plants per population) with EP3 and Minghui-86 rice lines in the designated Biosafety Assessment Centers in Fuzhou, Fujian Province. We selfed the F1 transgenic hybrids (EP3 × WR) to produce F2 and F3 hybrid lineages that segregated for the presence and absence of the target transgene to estimate the fitness effect of epsps transgene under the same genetic background in advanced generations of crop-wild hybrid descendants. The F2 transgenic hybrid plants used in the field experiment contained either hemizygous or homozygous epsps transgene, whereas the F3 transgenic hybrid plants only contained homozygous epsps transgene (Fig. 6).
Figure 6.
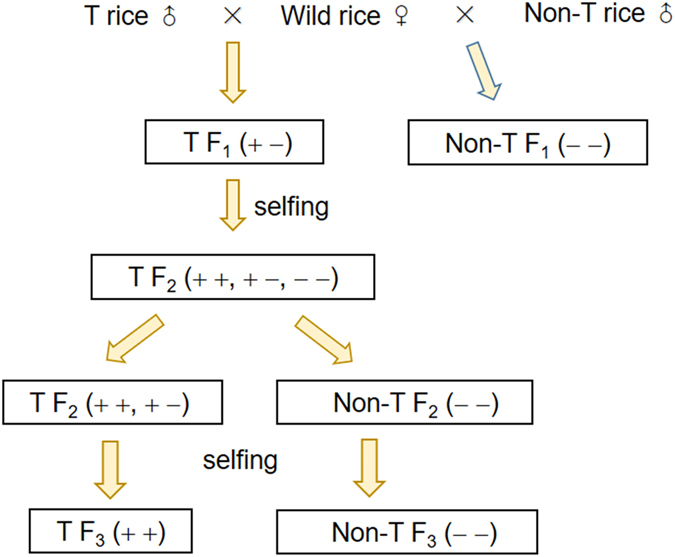
Schematic illustration of the pedigrees to produce F1-F3 crop-wild hybrid lineages. T: transgenic; + +, + −, and − −: transgene homozygous, transgene heterozygous, and non-transgenic, respectively. F1 and F2 hybrid lineages were used to test differences in fitness; the F3 hybrid lineages were used to test differences in seed germination after being buried in soils.
The identification of transgene status of crop-weed hybrid descendants in F1 and F2 generations was achieved by specific molecular markers16. For the F3 hybrids, we obtained transgene homozygous and non-transgene homozygous F3 hybrid lineages by randomly screening more than 15 seeds harvested from each F2 plant for the presence or absence of transgene. The above homozygous lineages were preserved for future F3 experiment. Thus, the plant materials used in the experiments included WR parents (WR1, WR2), F1 hybrids (WR1-F1+ and WR2-F1+ with presence of epsps transgene; WR1-F1- and WR2-F1- with absence of epsps transgene), F2 hybrid lineages (WR1-F2+ and WR2-F2+ with presence of epsps transgene; WR1-F2- and WR2-F2- with absence of epsps transgene), and F3 hybrid lineages (WR1-F3+ and WR2-F3+ with presence of epsps transgene; WR1-F3- and WR2-F3- with absence of epsps transgene).
Field experiment design
Common garden field experiments were carried out in the designated Biosafety Assessment Centers in Fuzhou, Fujian Province, to estimate the effects of epsps transgene on vegetative growth, phenology, fecundity, and overwintering regeneration. Six sets of materials were included in the experiments: transgenic F1 or F2 hybrid lineages and their non-transgenic F1 or F2 counterparts derived from WR1 and WR2, as well as the two wild parents. Two cultivation modes, pure planting of transgenic, non-transgenic hybrid lineages, or the parents and mix planting of transgenic and non-transgenic hybrid lineages alternately at different densities, were designed to estimate the competitive abilities between the transgenic and non-transgenic hybrid lineages. For each treatment, six replicates (plots) were included. In pure planting, each plot included 36 plants in 6 × 6 grid with 50 cm spacing for the F1 and F2 experiments. In mix planting, each plot also included 36 plants in 6 × 6 grid but with 30 cm spacing for the F1 and F2 experiments, and 40 cm and 50 cm spacing only for the F1 experiment. Consequently, a total of 72 (in F1) or 48 (in F2) plots were included for the field experiments. The field layout of all experimental plots followed a complete randomized design.
Seed burial experiments were carried out in the confined experimental blocks of Fudan University campus in Shanghai to estimate the germination ability of hybrid seeds after being buried in soils. Four groups of F3 hybrid seeds were included for the seed burial experiments: WR1-F3+, WR1-F3-, WR2-F3+ and WR2-F3-. These hybrid seeds were treated at 50 °C for 7 days to break the seed dormancy. The treated seeds were buried in the soil of a rice field after rice harvesting, for 0, 20, 40, and 60 days before seed germination. Consequently, a total of 16 treatments with three replicates (bags) and 48 bags were included in the experiments. Each nylon bag contained 50 seeds that was randomly buried in 10 cm depth of soils from December to next-year February. The buried seeds were moved out at the different days after burial and germinated on the moist filter papers in a petri dish at 30 °C to examine the seed germination (see detail in Table S5).
Correlation between endogenous EPSPS contents in parental plants and fitness change caused by the transgene
To study relationships between endogenous EPSPS protein contents and fitness changes, we measure EPSPS contents in transgene recipient parents, including WR (WR1 and WR2), WDR (WDR1 and WDR2), and cultivated rice (Minghui-86), using ELISA (enzyme linked immunosorbent assay), in addition to the increased panicles and seeds per plant. The EPSPS protein content was measured as the ratio (%) between the amount of EPSPS protein and the total amount of soluble proteins. Pooled leaf tissues from three plants were collected as one sample (replicate) and nine samples from each type of parental plants were included at the vegetative (60 days), reproductive (100 days), and ripening stages (160 days). The Quantiplate kit (Envirologix, Portland, OR, USA) was used for the detection of the EPSPS proteins following the ELISA manufacturer’s protocols. We set the wavelength of the microtiter plate reader to 450 nanometers (nm) using Plate Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The MICROPLATE MANAGER (MPM) software ver. 6 (Bio-Rad Laboratories, Inc.) was used to summarize the results. The ratios of increased number of panicles and seeds per plant were estimated between transgenic and non-transgenic crop-wild or crop-weed hybrid lineages in the F2 generation, and between EP3 and Minghui-83. Data of crop-weed hybrid lineages used in this study were from Wang et al. (2014).
Data collection and analysis
The methods for data collection follows the description in Table S5. Two-way ANOVAs were carried out to analyze the effects of transgene (transgenic vs. non-transgenic), wild parent (WR1 vs. WR2), and their interaction on fitness in pure-planting plots. Independent and paired t-tests were used to determine differences between transgenic and non-transgenic hybrid lineages for fitness-related traits in pure-planting pots and mix-planting plots, respectively. Independent t-tests was used to detect differences in endogenous EPSPS contents between WR1 and WR2 based on the ELISA experiment. The correlation between endogenous EPSPS protein contents and fitness changes was calculated based on Pearson Correlation Coefficient. All statistical analyses were performed using the software IBM SPSS Statistics ver. 22.0 for Windows (SPSS Inc., IBM Company Chicago, IL, USA, 2010).
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank Allison Snow for the suggestion of this paper. This work is supported by the Natural Science Foundation of China (#31330014) and the National Program of Development of Transgenic New Species of China (2017ZX08011-006).
Author Contributions
B.-R.L., X.Y., J.S. and F.W. conceived and designed the experiment. X.Y., L.L., X.J. C.X. and W.W. performed field experiment, X.Y and B.-R.L. analyzed the data and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07089-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ellstrand NC, Prentice HC, Hancock JF. Gene flow and introgression from domesticated plants into their wild relatives. Annu Rev Ecol Syst. 1999;30:539–563. doi: 10.1146/annurev.ecolsys.30.1.539. [DOI] [Google Scholar]
- 2.Snow AA, et al. Genetically engineered organisms and the environment: current status and recommendations. Ecol Appl. 2005;15:377–404. doi: 10.1890/04-0539. [DOI] [Google Scholar]
- 3.Lu B-R, Yang C. Gene flow from genetically modified rice to its wild relatives: assessing potential ecological consequences. Biotechnol Adv. 2009;27:1083–1091. doi: 10.1016/j.biotechadv.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Ellstrand NC. Current knowledge of gene flow in plants: implications for transgene flow. Philos Trans R Soc Lond B Biol Sci. 2003;358:1163–1170. doi: 10.1098/rstb.2003.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu B-R, Snow AA. Gene flow from genetically modified rice and its environmental consequences. BioScience. 2005;55:669–678. doi: 10.1641/0006-3568(2005)055[0669:GFFGMR]2.0.CO;2. [DOI] [Google Scholar]
- 6.Ellstrand NC, et al. Introgression of crop alleles into wild or weedy populations. Annu Rev Ecol Evol Syst. 2013;44:325–345. doi: 10.1146/annurev-ecolsys-110512-135840. [DOI] [Google Scholar]
- 7.Lu B-R, Yang X, Ellstrand NC. Fitness correlates of crop transgene flow into weedy populations: a case study of weedy rice in China and other examples. Evol Appl. 2016;9:857–870. doi: 10.1111/eva.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellstrand NC, Rieseberg LH. When gene flow really matters: gene flow in applied evolutionary biology. Evol Appl. 2016;9:833–836. doi: 10.1111/eva.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasu M, Ferrari MJ, Stephenson AG. Interrelationships among a virus-resistance transgene, herbivory, and a bacterial disease in a wild Cucurbita. Int J Plant Sci. 2010;171:1048–1058. doi: 10.1086/656531. [DOI] [Google Scholar]
- 10.Sasu M, Ferrari MJ, Du D, Winsor JA, Stephenson AG. Indirect costs of a nontarget pathogen mitigate the direct benefits of a virus-resistant transgene in wild Cucurbita. Proc Natl Acad Sci U S A. 2009;106:19067–19071. doi: 10.1073/pnas.0905106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guadagnuolo R, Clegg J, Ellstrand NC. Relative fitness of transgenic vs. non-transgenic maize x teosinte hybrids, a field evaluation. Ecol Appl. 2006;16:1967–1974. doi: 10.1890/1051-0761(2006)016[1967:RFOTVN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Burke JM, Rieseberg LH. Fitness of transgenic disease resistance in sunflower. Science. 2003;300:1250. doi: 10.1126/science.1084960. [DOI] [PubMed] [Google Scholar]
- 13.Yang, X., Wang, F., Su, J. & Lu, B.-R. Limited fitness advantages of crop-weed hybrid progeny containing insect-resistant transgenes (Bt/CpTI) in transgenic rice field. PLoS ONE7, e41220 (2012). [DOI] [PMC free article] [PubMed]
- 14.Yang X, et al. Efficacy of insect-resistance Bt/CpTI transgenes in F5–F7 generations of rice crop–weed hybrid progeny: implications for assessing ecological impact of transgene flow. Sci Bull. 2015;60:1563–1571. doi: 10.1007/s11434-015-0885-x. [DOI] [Google Scholar]
- 15.Yang X, et al. Transgenes for insect resistance reduce herbivory and enhance fecundity in advanced generations of crop-weed hybrids of rice. Evol Appl. 2011;4:672–684. doi: 10.1111/j.1752-4571.2011.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. A novel 5‐enolpyruvoylshikimate‐3‐phosphate (EPSP) synthase transgene for glyphosate resistance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide. New Phytol. 2014;202:679–688. doi: 10.1111/nph.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, et al. Limited ecological risk of insect-resistance transgene flow from cultivated rice to its wild ancestor based on life-cycle fitness assessment. Sci Bull. 2016;61:1440–1450. doi: 10.1007/s11434-016-1152-5. [DOI] [Google Scholar]
- 18.Xia H, et al. Ambient insect pressure and recipient genotypes determine fecundity of transgenic crop‐weed rice hybrid progeny: Implications for environmental biosafety assessment. Evol Appl. 2016;9:847–856. doi: 10.1111/eva.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green JM. The benefits of herbicide‐resistant crops. Pest Manag Sci. 2012;68:1323–1331. doi: 10.1002/ps.3374. [DOI] [PubMed] [Google Scholar]
- 20.Duke SO. Perspectives on transgenic, herbicide‐resistant crops in the United States almost 20 years after introduction. Pest Manag Sci. 2015;71:652–657. doi: 10.1002/ps.3863. [DOI] [PubMed] [Google Scholar]
- 21.Gen, A. GM CROPS. Nature 497 (2013).
- 22.Rao AN, Johnson DE, Sivaprasad B, Ladha JK, Mortimer AM. Weed management in direct-seeded rice. Adv Agron. 2007;93:153–255. doi: 10.1016/S0065-2113(06)93004-1. [DOI] [Google Scholar]
- 23.Ziska, L. H. et al. Chapter Three-Weedy (Red) Rice: An emerging constraint to global rice production. Adv Agron129, 181–228 (2015).
- 24.Cao Q, et al. Genetic diversity and origin of weedy rice (Oryza sativa f. spontanea) populations found in north-eastern China revealed by simple sequence repeat (SSR) markers. Ann Bot. 2006;98:1241–1252. doi: 10.1093/aob/mcl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia HB, Wang W, Xia H, Zhao W, Lu B-R. Conspecific crop-weed introgression influences evolution of weedy rice (Oryza sativa f. spontanea) across a geographical range. PLoS ONE. 2011;6:e16189. doi: 10.1371/journal.pone.0016189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harden J, et al. ProvisiaTM: a new vision in red rice control. Weed Sci Am Abstr. 2014;54:198. [Google Scholar]
- 27.Shivrain VK, et al. Gene flow between Clearfield™ rice and red rice. Crop Prot. 2007;26:349–356. doi: 10.1016/j.cropro.2005.09.019. [DOI] [Google Scholar]
- 28.Merotto A, et al. Evolutionary and social consequences of introgression of nontransgenic herbicide resistance from rice to weedy rice in Brazil. Evol Appl. 2016;9:837–846. doi: 10.1111/eva.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichman JR, et al. Establishment of transgenic herbicide‐resistant creeping bentgrass (Agrostis stolonifera L.) in nonagronomic habitats. Mol Ecol. 2006;15:4243–4255. doi: 10.1111/j.1365-294X.2006.03072.x. [DOI] [PubMed] [Google Scholar]
- 30.Warwick SI, Legere A, Simard MJ, James T. Do escaped transgenes persist in nature? The case of an herbicide resistance transgene in a weedy Brassica rapa population. Mol Ecol. 2008;17:1387–1395. doi: 10.1111/j.1365-294X.2007.03567.x. [DOI] [PubMed] [Google Scholar]
- 31.Bergelson J, Purrington CB, Palm CJ, LOpez-Gutierrez JC. Costs of resistance: a test using transgenic Arabidopsis thaliana. P Roy Soc B-Biol. 1996;263:1659–1663. doi: 10.1098/rspb.1996.0242. [DOI] [PubMed] [Google Scholar]
- 32.Snow AA, Andersen B, Jørgensen RB. Costs of transgenic herbicide resistance introgressed from Brassica napus into weedy B. rapa. Mol Ecol. 1999;8:605–615. doi: 10.1046/j.1365-294x.1999.00596.x. [DOI] [Google Scholar]
- 33.Vila-Aiub MM, Gundel PE, Preston C. Experimental methods for estimation of plant fitness costs associated with herbicide resistance genes. Weed Sci. 2015;63(sp1):203–216. doi: 10.1614/WS-D-14-00062.1. [DOI] [Google Scholar]
- 34.Su, J., Chen, G. M., Tian, D. G., Zhu, Z. & Wang, F. A gene encodes 5-enolpyruvylshikimate-3-phosphate mutagenized by error-prone PCR conferred rice with high glyphosate-tolerance. Mol Plant Breed6, 830–836 (2008) (in Chinese with English Abstract)
- 35.Zhang WX, Yang QW. Collecting, Evaluation and conservation of wild rice resources in China. J Plant Genet Resour. 2003;4:369–373. [Google Scholar]
- 36.Xie Z, Lu Y, Ge S, Hong D, Li F. Clonality in wild rice (Oryza rufipogon, Poaceae) and its implications for conservation management. Am J Bot. 2001;88:1058–1064. doi: 10.2307/2657088. [DOI] [PubMed] [Google Scholar]
- 37.Vaughan DA, Lu B-R, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. doi: 10.1016/j.plantsci.2008.01.016. [DOI] [Google Scholar]
- 38.Lin, S. C. & Yuan, L. P. Hybrid rice breeding in China. In: Innovative approaches in Rice Breeding, pp. 35–51, Int. Rice Res. Inst. Manila, Philippines (1980).
- 39.Xiao. J, et al. A wild species contains genes that may significantly increase the yield of rice. Nature. 1996;384:223–224. doi: 10.1038/384223a0. [DOI] [Google Scholar]
- 40.Bellon, M. R., Brar, D. S., Lu, B.-R. & Pham, J. L. Rice genetic resources. In: Dwoling NG, Greenfield SM, Fischer KS (eds) Sustainability of rice in the global food system. Chapter 16, Davis, California (USA). Pacific Basin Study Center and IRRI, Manila, pp 251–283 (1998).
- 41.Zhao Y, et al. Are habitat fragmentation, local adaptation and isolation‐by‐distance driving population divergence in wild rice Oryza rufipogon? Mol Ecol. 2013;22:5531–5547. doi: 10.1111/mec.12517. [DOI] [PubMed] [Google Scholar]
- 42.Gao LZ, Gao CW. Lowered diversity and increased inbreeding depression within peripheral populations of wild rice Oryza rufipogon. PLoS ONE. 2016;11:e0150468. doi: 10.1371/journal.pone.0150468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu B-R. Introgression of transgenic crop alleles: Its evolutionary impacts on conserving genetic diversity of crop wild relatives. J Syst Evol. 2013;51:245–262. doi: 10.1111/jse.12011. [DOI] [Google Scholar]
- 44.Song ZP, Lu B-R, Zhu YG, Chen JK. Gene flow from cultivated rice to the wild species Oryza rufipogon under experimental field conditions. New Phytol. 2003;157:657–665. doi: 10.1046/j.1469-8137.2003.00699.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang F, et al. A large-scale field study of transgene flow from cultivated rice (Oryza sativa) to common wild rice (O. rufipogon) and barnyard grass (Echinochloa crusgalli) Plant Biotechnol J. 2006;4:667–676. doi: 10.1111/j.1467-7652.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 46.Song ZP, et al. Evidences of introgression from cultivated rice to Oryza rufipogon (Poaceae) populations based on SSR fingerprinting: implications for wild rice differentiation and conservation. Evol Ecol. 2006;20:501–522. doi: 10.1007/s10682-006-9113-0. [DOI] [Google Scholar]
- 47.Giacomini D, Westra P, Ward SM. Impact of genetic background in fitness cost studies: An example from glyphosate-resistant palmer amaranth. Weed Sci. 2014;62:29–37. doi: 10.1614/WS-D-13-00066.1. [DOI] [Google Scholar]
- 48.Kumar V, Jha P. Growth and reproduction of glyphosate-resistant and susceptible populations of Kochia scoparia. PLoS ONE. 2015;10:e0147779. doi: 10.1371/journal.pone.0142675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vila-Aiub MM, et al. No fitness cost of glyphosate resistance endowed by massive EPSPS gene amplification in Amaranthus palmeri. Planta. 2014;239:793–801. doi: 10.1007/s00425-013-2022-x. [DOI] [PubMed] [Google Scholar]
- 50.Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic compounds. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franz, J. E., Mao, M. K. & Sikorski, J. A. Glyphosate: a unique global herbicide. American Chemistry Society, Washington, DC, USA. (1997).
- 52.Yang X, et al. Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana. PLoS ONE. 2017;12:e0175820. doi: 10.1371/journal.pone.0175820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- 54.Tohge T, Watanabe M, Hoefgen R, Fernie AR. Shikimate and phenylalanine biosynthesis in the green lineage. Front Plant Sci. 2013;4:62. doi: 10.3389/fpls.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Owen MDK, et al. Comparisons of genetically modified and non-genetically modified soybean cultivars and weed management systems. Crop Sci. 2010;50:2597–2604. doi: 10.2135/cropsci2010.01.0035. [DOI] [Google Scholar]
- 56.Zhou H, et al. Field efficacy assessment of transgenic Roundup Ready wheat. Crop Sci. 2003;43:1072–1075. doi: 10.2135/cropsci2003.1072. [DOI] [Google Scholar]
- 57.Vaughan, D. A. The wild relatives of rice: a genetic resources handbook. Int. Rice Res. Inst. (1994).
- 58.Ellstrand NC. Is gene flow the most important evolutionary force in plants? Am J Bot. 2014;101:737–753. doi: 10.3732/ajb.1400024. [DOI] [PubMed] [Google Scholar]
- 59.Xu JW, Feng DJ, Li XG, Chang TJ, Zhu Z. Cloning of genomic DNA of rice 5-enolpyruvylshikimate 3-phosphate synthase gene and chromosomal localization of the gene. Sci Sin. 2002;45:251–259. doi: 10.1360/02yc9028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


