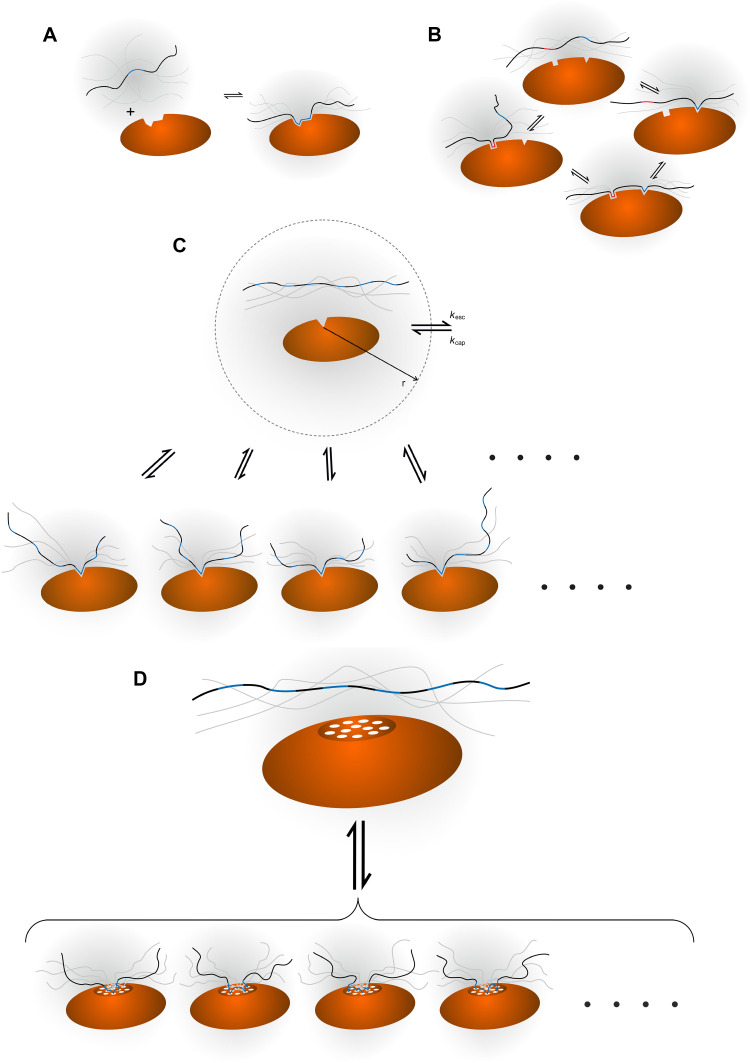
Fig. 1.
Illustration of four different binding mechanisms involving IDPs. Different ligand-receptor interactions involving an IDP ligand (wavy black string) and a macromolecular receptor (orange oval) are shown. The binding epitopes on the ligand are highlighted in blue or (red), whereas a binding site on the receptor is symbolised with an indentation. a Simple two-state binding, implying the existence of either the free or the bound state with no intermediates. There is a linear correlation between the concentration of ligand and the fraction of molecules in the bound state. b Avidity, where two or more epitopes on the ligand and a corresponding number of binding sites on the receptor will interact. If one interaction is established, a second binding event is more likely to occur due to the proximity of the additional interaction site(s). c Allovalency. A single binding site on the receptor can bind to several, identical epitopes along the ligand. When a bound ligand is released, the chance of rebinding is higher than anticipated from the ligand concentration alone, because of the very high epitope concentration close to the binding site on the receptor. An illustration of the capture sphere is shown with the corresponding rate constants. d Fuzzy complex. Both molecules have a number of interaction sites (shown as white circles on the receptor). An interaction site on the ligand is not restricted to connect with one specific interaction site on the receptor and vice versa. Thus, when one interaction is lost, the probability of forming another one because of the high local concentration of both ligand and receptor binding sites is much higher than what one would anticipate from ligand concentration alone. The smaller representation of the bound states in c and d is intentional, but not real, and is drawn for clarity only