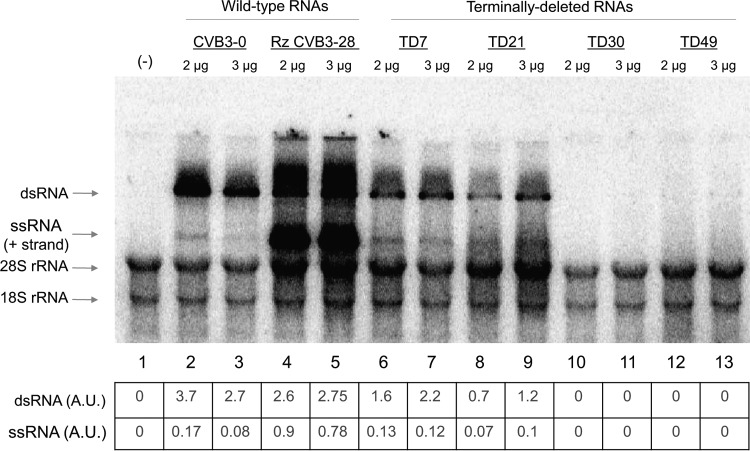
FIG 5.
In vitro RNA replication of wild-type and TD coxsackievirus B3 transcripts. After a 4.5-h in vitro translation assay allowing the synthesis of nonstructural proteins in the absence of [35S]methionine, the reaction mixture, consisting of viral transcripts (2 μg or 3 μg) and HeLa S10 cytoplasmic extract, was incubated for an additional 2 h at 34°C for RNA synthesis in the presence of [α-32P]CTP. The purified RNA was then resolved on a 1.1% agarose gel in Tris-borate-EDTA buffer containing ethidium bromide. The amounts of 18S and 28S rRNA in each lane were used to confirm equal loading of samples in the gel. Plasmid DNA template encoding CVB3-28 RNA harbors a hammerhead ribozyme at the 5′ end of the transcript (32), producing an authentic CVB3 5′ terminus. This RNA is an efficient template for negative-strand RNA synthesis and subsequent rounds of positive-strand RNA synthesis. In vitro RNAs produced from pCVB3-0 have no hammerhead ribozyme at the 5′ end and harbor nonviral sequences that result in efficient negative-strand RNA synthesis but very inefficient positive-strand RNA (ssRNA) synthesis (13, 35). All TD transcripts harbor a hammerhead ribozyme at their 5′ termini, producing authentic 5′-terminal CVB3 sequences. The signals detected in the gel for single-stranded and double-stranded viral RNAs were quantified using ImageJ software. The signals were normalized to the 28S and 18S rRNA levels (collectively) for each sample and expressed in arbitrary units (A.U.). Lane 1 (−) shows the results of incubation of the in vitro replication reaction mixture in the absence of transcript RNA.