Abstract
Background
Hypoxia Inducible Factor 3 Alpha Subunit (HIF3A) DNA has been demonstrated to be associated with obesity in the methylation level, and it also has a Body Mass Index (BMI)-independent association with plasma alanine aminotransferase (ALT). However, the relation among obesity, plasma ALT, HIF3A polymorphism and methylation remains unclear. This study aims to identify the association between HIF3A polymorphism and plasma ALT, and further to determine whether the effect of HIF3A polymorphism on ALT could be modified by obesity or mediated by DNA methylation.
Methods
The HIF3A rs3826795 polymorphism was genotyped in a case-control study including 2030 Chinese children aged 7–18 years (705 obese cases and 1325 non-obese controls). Furthermore, the HIF3A DNA methylation of the peripheral blood was measured in 110 severely obese children and 110 age- and gender- matched normal-weight controls.
Results
There was no overall association between the HIF3A rs3826795 polymorphism and ALT. A significant interaction between obesity and rs3826795 in relation with ALT was found (P inter = 0.042), with rs3826795 G-allele number elevating ALT significantly only in obese children (β’ = 0.075, P = 0.037), but not in non-obese children (β’ = −0.009, P = 0.741). Additionally, a mediation effect of HIF3A methylation was found in the association between the HIF3A rs3826795 polymorphism and ALT among obese children (β’ = 0.242, P = 0.014).
Conclusion
This is the first study to report the interaction between obesity and HIF3A gene in relation with ALT, and also to reveal a mediation effect among the HIF3A polymorphism, methylation and ALT. This study provides new evidence to the function of HIF3A gene, which would be helpful for future risk assessment and personalized treatment of liver diseases.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-017-0437-0) contains supplementary material, which is available to authorized users.
Keywords: Hypoxia inducible factor 3 alpha subunit, Alanine aminotransferase, Single nucleotide polymorphisms, DNA Methylation, Obesity, Children
Background
The prevalence of childhood obesity has witnessed a constant increase worldwide. In the year of 2013, 23·8% of boys and 22·6% of girls were overweight or obese in developed countries, and 12.9% of boys and 13.4% of girls were overweight or obese in developing countries [1].
Obesity could raise the risk of multiple co-morbid complications, including the Non-Alcoholic Fatty Liver Disease (NAFLD), type 2 diabetes, hypertension, cardiovascular disease, stroke and several kinds of cancers [2, 3]. It has been demonstrated by a meta-analysis study that the prevalence of childhood NAFLD is 7.6% in the general population, and could increase to 34.2% among obese children [4]. Usually used as a biomarker to reflect liver function clinically, the plasma alanine aminotransferase (ALT) is also reported to be associated with higher weight, body mass index (BMI) and waist circumference [5].
Evidence suggested that there could be a strong genetic background in the predisposition to both obesity [6] and NAFLD [7]. Meanwhile, epigenetics (heritable events not caused by changes in DNA sequence) could also contribute to the development of obesity [8] and NAFLD [9]. Being the most stable epigenetic marker, DNA methylation can not only be affected by the environment, but also modulated by genetic variants. It is reported that DNA methylation correlates with nearby Single Nucleotide Polymorphisms (SNPs), and studies conducted in various kinds of tissues (including brain, adipose, blood, et al.) have revealed that there are quantitative trait loci (QTLs) for DNA methylation, also called methylation QTLs (meQTLs) [10–12].
Previous studies have shed light on the association between Hypoxia Inducible Factor 3 Alpha Subunit (HIF3A) DNA methylation and obesity-related traits. Dick et al. [13] conducted an Epigenome-Wide Association Study (EWAS) among European white adults, uncovering a specific association between BMI and DNA methylation at 3 CpG (Cytosine-Phosphate-Guanine dinucleotides) sites of the HIF3A gene, and also reported nearby SNPs including the HIF3A rs3826795 polymorphism to be associated with methylation. In our previous study [14], we have reported that obese children had a relatively higher level of HIF3A DNA methylation, and HIF3A methylation had a BMI-independent association with plasma ALT. However, the relation among obesity, plasma ALT, HIF3A SNPs and methylation remains unclear.
In this paper, genotyping for the HIF3A rs3826795 polymorphism among 2030 Chinese children aged 7–18 years old was performed, and the HIF3A methylation data of 220 children detected in our previous study [14] was also used. The aims of this study are: (1) identifying the association between the HIF3A rs3826795 polymorphism and plasma ALT; (2) determining whether obesity could interact with rs3826795 on plasma ALT; and (3) investigating whether the effect of rs3826795 on ALT is mediated by HIF3A DNA methylation.
Methods
Subjects
We conducted a case-control study among 2030 Chinese children aged 7–18 years old from two independent study groups, including 705 obese cases and 1325 non-obese controls recruited from the urban regions of Beijing, China. The first study group came from the study on Adolescent Lipids, Insulin Resistance, and candidate genes (ALIR). The second study group was from the Comprehensive Prevention project for Overweight and Obese Adolescents (CPOOA). All obese individuals in the selected schools were recruited with their voluntary participation. The method of cluster sampling was adopted to recruit non-obese subjects from some classes of each grade in the same schools. The ALIR subjects were ascertained from adolescents aged 14–17 years in nine middle schools of Dongcheng District of Beijing, including 386 obese adolescents and 551 non-obese adolescents. The CPOOA subjects were recruited from children and adolescents aged 7–18 years old in five elementary and middle schools of the Haidian District of Beijing, comprising 319 obese children and adolescents and 774 non-obese children and adolescents. The ascertainment strategies for the two study groups have been described in detail previously [15, 16]. We used the uniform BMI percentile criteria for obese and non-obese children, which were determined in a representative Chinese population [17]. According to the criteria, the children and adolescents with an age- and gender-specific BMI ≥ 95th percentile are defined as obese, whereas those with a BMI between 15th and 95th percentile are non-obese. The individuals with any cardiovascular or metabolic disease were excluded. Anthropometric measurements, including height and weight, were measured at school according to standard protocols. Fasting venous blood samples were taken for detection of ALT.
Methylation data were collected from 110 severely obese children and 110 normal-weight age- and gender-matched controls, which were chosen from the CPOOA study. We chose those with an age- and gender-specific BMI ≥ 97th percentile as the severely obese cases, and those with BMI between 15th and 85th percentile as non-obese controls. We point the reader to our prior work, where we have described the study design for the methylation detection [14].
Studies were approved by the Ethic committee of Peking University Health Science Center. Written informed consent was provided by all participants and, in the case of minors, by their parents.
SNP genotyping
The HIF3A rs3826795 polymorphism was genotyped using genomic DNAs extracted from blood leukocytes by the phenol-chloroform extraction method. Genotyping was conducted on MassARRAY System (Sequenom, San Diego, CA, USA). Primers, including a pair of amplification primers and an extension primer, were designed with Sequenom MassArray Assay Design Suite. A multiplex polymerase chain reaction was performed, and unincorporated double stranded nucleotide triphosphate bases were dephosphorylated with shrimp alkaline phosphatase followed by primer extension. The purified primer extension reaction was spotted on to a 384-element silicon chip (SpectroCHIP, Sequenom) and analyzed in the Matrix assisted laser desorption ionization time of flight mass Spectrometry (MALDI-TOF MS, Sequenom). The resulting spectra were processed with MassArray Typer (Sequenom) (http://www.sequenom.com). The genotyping call rate of the HIF3A rs3826795 polymorphism was 97.9%. All the experiments were done by investigators who were blind to the phenotypes.
DNA methylation detection
Details of the methylation examination were described previously [14]. The HIF3A DNA methylation data were collected from genomic DNA of peripheral blood leukocytes among 110 obese children and 110 non-obese controls. The blood samples were the same for SNP genotyping and methylation detection. We used Sequenom’s MassARRAY system (Sequenom, San Diego, CA) to perform quantitative methylation analyses [18]. A fully-methylated positive control and a 0-methylated negative control were included for each run. Methylation detection was conducted in duplicate for all the samples. Three samples failed to give a reliable PCR product, and 5 samples were excluded from analysis samples because of low call rates (<80%). Thus, we obtained data of 9 CpG sites (located form 46,801,557 to 46,801,760) of HIF3A DNA methylation among 107 obese children and 105 controls.
Statistical analyses
Quanto software (University of Southern California, Los Angeles, CA) was used to conduct power analysis. As no study has previously reported the interaction between obesity and the HIF3A rs3826795 polymorphism on ALT, we could only estimate the sample size. Using the additive genetic model, at a two-sided significance level of P < 0.05, with effect allele frequency of 0.41 (referring to the risk allele frequency of Singapore population [19]), with the proportion of obese children as 0.35 [20], assuming the effects on logALT of gene, obesity and gene × obesity were 0.10, 0.30 and 0.20 respectively, 1906 people were estimated to obtain a statistical power of 85%.
The genotype data were tested for deviation from Hardy-Weinberg equilibrium. The polymorphism was analyzed under additive models. For the values of ALT, we used the log converted values (logALT) because of the skewed distribution of the original values, and then we used the criterion of Mean ± 3SD and excluded 26 extreme values. Differences in general characteristics between obese and non-obese subjects were tested with Chi-square (categorical variables) or t-test (continuous variables). Linear regression models adjusting for age, age 2 and gender were used to examine the effect of SNP on logALT. In order to fully control the effect of age on ALT, age and age2 were adjusted for simultaneously, since phenotypes could be highly influenced by age especially in the children population, and the association between the phenotype and age could be non-linear. The method of age-squared adjustment was also used in previous studies [21, 22]. Interaction analyses were conducted using stratified analyses and the statistical tests for the interaction term.
The mediation analysis was based on the model brought forward by Baron, et al. [23], and was used by other literatures, especially in the domain of psychology [24]. Three multivariate linear regression models were conducted, all adjusting for age, age2 and gender: (1) Y = cX + p 1 age + q 1 age 2 + r 1 gender + e 1; (2) M = ax + p 2 age + q 2 age 2 + r 2 gender + e 2; (3) Y = c r X + bM + p 3 age + q 3 age 2 + r 3 gender + e 3. The statistical test of the mediation effect included several steps: (1) the association between the independent variable and the dependent variable was tested (the coefficient c); (2) the association between the independent variable and the potential mediator was tested (the coefficient a); (3) both of the independent variable and the potential mediator were entered simultaneously as predictors of the dependent variable, and the coefficient b was tested to establish the significance of the mediation effect; (4) if the mediation effect was significant, the type of mediation effect could be determined by testing the coefficient c r, indicating either full mediation effect (c r not significant) or partial mediation effect (c r still significant).
A two-sided P < 0.05 was considered as nominally significant. Statistical analyses were performed using SPSS 18.0 software (SPSS Inc., Chicago, IL).
Results
General characteristics of the study groups
General characteristics of obese cases and non-obese controls are summarized in Table 1 (Dataset in Additional file 1). The age of the participants ranged from 7 to 18 years old, with the average age of 12.9 ± 2.7 years old. We found no significant difference in age between obese and non-obese groups (P > 0.05), but there was a higher proportion of boys in the obese group than in the non-obese group (P < 0.001). Compared to non-obese children, obese children had higher plasma ALT levels (P < 0.001). The difference of ALT between obese and non-obese groups remained significant after adjusting for age, age 2 and gender (P < 0.001). Details of the characteristics of the 110 extremely obese children and 110 matched controls with methylation examination were described previously [14]. Briefly, there was no difference between the two groups in age (P = 0.934) or gender (P = 0.946), and the 110 extremely obese children had a higher level of ALT as compared with controls (P < 0.001).
Table 1.
General characteristics of the study groups
| Total | Obese | Non-obese | P | |
|---|---|---|---|---|
| Number | 2030 | 705 | 1325 | |
| Boys (%) | 1218(60.0) | 484(68.7) | 734(55.4) | <0.001 |
| Age (year) | 12.9 ± 2.7 | 12.8 ± 2.6 | 12.9 ± 2.7 | 0.497 |
| Body mass index (BMI, kg/m2) | 23.8 ± 4.8 | 28.1 ± 3.9 | 21.5 ± 3.5 | <0.001 |
| Alanine aminotransferase (ALT, IU/L) | 1.12 ± 0.27 | 1.26 ± 0.29 | 1.05 ± 0.24 | <0.001 |
Values are provided as Mean ± SD if not indicated otherwise
For ALT, the log converted values are used instead of the original values
Interaction analyses of obesity and the HIF3A rs3826795 polymorphism on plasma ALT levels
The risk allele (G allele) frequency of the HIF3A rs3826795 polymorphism was 48.3% in the current study. The genotype distribution of polymorphism in the control group was in Hardy-Weinberg equilibrium (P > 0.05). We first examined the association between the variant and obesity, but found that rs3826795 was not significantly associated with obesity (P = 0.843), or BMI (P = 0.268) (both adjusting for age, age2 and gender). We then divided the study population by rs3826795 genotype and obesity group (obese vs non-obese), and compared the ALT levels in different subgroups (Table 2). The association between ALT and the HIF3A rs3826795 genotype was not significant in the overall population (P = 0.340). By conducting stratified analyses, we found that the number of HIF3A rs3826795 G-allele was positively associated with elevated plasma ALT in obese children (β’ = 0.075, P = 0.037), with each G-allele increasing the logALT level by 0.030 (which was the unstandardized coefficient of SNP in the regression model). We observed no significant association between ALT and the HIF3A rs3826795 genotype in non-obese children (P = 0.741). Using a multivariate linear regression model, we then tested the interaction term (SNP × obesity group), and found a significant interaction between G-allele number in the HIF3A rs3826795 polymorphism and obesity on the plasma ALT (P = 0.042).
Table 2.
Interaction analyses of the HIF3A rs3826795 polymorphism and obesity on plasma ALT
| Mean ± SD of logALT (IU/L) | ||||||
|---|---|---|---|---|---|---|
| AA (n = 535) | AG (n = 985) | GG (n = 468) | β’ | P | P inter | |
| Total | 1.11 ± 0.27 | 1.12 ± 0.27 | 1.14 ± 0.28 | 0.020 | 0.340 | |
| Non-obese | 1.04 ± 0.25 | 1.06 ± 0.23 | 1.05 ± 0.23 | −0.009 | 0.741 | 0.042 |
| Obese | 1.24 ± 0.26 | 1.25 ± 0.30 | 1.30 ± 0.29 | 0.075 | 0.037 | |
β’ stands for the standardized coefficient of G allele number in the model
P inter stands for the P value of the interaction term in the linear regression model, including logALT as the dependent variable, and SNP, obesity group, SNP × obesity group, age, age2 and gender as independent variables
HIF3A: Hypoxia Inducible Factor 3 Alpha Subunit; ALT alanine aminotransferase
Mediation analysis of the HIF3A rs3826795 polymorphism, methylation and plasma ALT
HIF3A DNA methylation was tested in 110 children with severe obesity and 110 non-obese age- and gender- matched controls. We used the data of CpG unit at 46801699 to conduct the mediation analysis among HIF3A SNP, methylation and ALT.
We used a mediation model to examine whether the HIF3A methylation acted as a mediator in the association between the HIF3A rs3826795 polymorphism and plasma ALT, as shown in Fig. 1. Here ‘a’, ‘b’, ‘c’ and ‘c r ’ were used to represent the coefficients in the mediation analysis model. In the 110 obese children with methylation data, the HIF3A rs3826795 G-allele number was positively associated with elevated ALT (c = 0.203, P = 0.034), with each G-allele increasing the logALT level by 0.085. There was a positive association between the HIF3A rs3826795 G-allele number and HIF3A methylation (a = 0.252, P = 0.009), with each G-allele increasing the methylation level by 0.032. By testing ‘b’, we found the mediation effect was significant (b = 0.242, P = 0.014), and the insignificance of ‘c r’ indicated a complete mediation effect (c’ = 0.140, P = 0.144), meaning that the effect between SNP and ALT in obese children would not be significant without methylation as the mediator. Further calculation (ab/c × 100%) reflected the mediation effect was 70.0%, meaning that 70.0% of the total effect between SNP and ALT was mediated by DNA methylation.
Fig. 1.
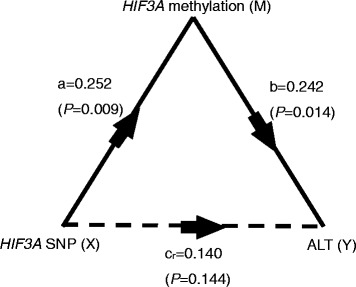
Mediation analysis in obese children for HIF3A SNP, methylation and plasma ALT. ALT: alanine aminotransferase; HIF3A: Hypoxia Inducible Factor 3 Alpha Subunit; SNP: Single Nucleotide Polymorphism. In order to distinguish among the three linear regression models constructed in the mediation analysis, the coefficients in the model were represented by ‘a’,’ b’, ‘c’ and ‘c r ’. The first regression model used ‘c’ as the coefficient of SNP in association with ALT; the second model used ‘a’ as the coefficient of SNP in association with methylation; the third model put methylation and SNP as independent variables simultaneously, and used ‘b’ and ‘c r ’ as the coefficients in association with ALT. ‘X’, ‘Y’ and ‘M’ were used to represent the independent variable, the dependent variable and the mediator in the mediation analysis model
Further, we conducted analyses in the 110 non-obese children, and found no significant association between any two variables among the HIF3A rs3826795 polymorphism, HIF3A methylation and ALT (P > 0.05). No association was observed between rs3826795 and the methylation of the CpG site at 46801699 (β’ = 0.198, P = 0.070), while there was no significant association between the HIF3A rs3826795 and ALT (β’ = 0.023, P = 0.829), or between HIF3A methylation and ALT (β’ = 0.146, P = 0.160).
Discussion
To the best of our knowledge, our study is the first to find that 1) the HIF3A rs3826795 polymorphism interacts with obesity on plasma ALT, with the rs3826795 G-allele number elevating the ALT level only in obese children; and 2) the effect of the HIF3A rs3826795 polymorphism on ALT in obese children is mediated by HIF3A DNA methylation.
HIF3A encodes for the protein of HIF-3α, which is the α-3 subunit of the hypoxia inducible transcription factors (HIFs). Uncovered in 1992, HIF acts as a heterodimeric transcription factor composing a β subunit (HIF-β) and one of three α subunits (HIF-1α, HIF-2α, HIF-3α), and regulates many adaptive responses to low oxygen tension (hypoxia) on both cellular and physiological levels [25]. HIF-3α has not been investigated as thoroughly as the other α subunits, but it is usually regarded as a negative regulator of HIF-1α and HIF-2α [26]. The HIF3A gene locates at 19q13.2 with a length of 43 kb, containing 19 introns and 8 kinds of alternative splicing [27].
The risk allele frequency of the HIF3A rs3826795 polymorphism was 0.79–0.82 in white adults of the EWAS study, and another study [28] focusing on HIF3A variant and weight change reported the risk allele frequency of rs3826795 to be 0.82–0.83 in US adults of European ancestry. Pan et al. [19] examined in Singapore mother–offspring pairs and reported the risk allele frequency of rs3826795 to be 0.41. In the current study we found that the risk allele frequency of rs3826795 was 0.48 in Chinese children, which is lower than that in the population of European ancestry, and similar to Singapore population.
The rs3826795 polymorphism and CpG site 46,801,699 both locate at the first intron of the HIF3A gene, with the CpG site about 1.3 kb downstream rs3826795. In the current study we found that the HIF3A rs3826795 polymorphism could affect the HIF3A methylation at CpG site 46,801,699 and other nearby CpG sites, indicating rs3826795 as a methylation quantitative-trait locus (metQTL). Known to be influenced by environmental factors, DNA methylation can also be modulated by genetic variants. Grundberg et al. [10] conducted analysis of metQTL in a genome study and revealed that 28% genome CpGs were associated with nearby SNPs (within 100 kb). The study further identified that meQTLs over-lapping metabolic disease loci were enriched in genetic enhancer. Another study conducted by Voisin et al. [12] also found that meQTLs tended to locate in intergenic regions, showing that obesity-associated SNPs were underrepresented in promoters but enriched in intergenic regions.
In the EWAS study, Dick et al. [13] reported the HIF3A rs3826795 polymorphism to be associated with methylation at a nearby CpG site (46801642), with each risk allele (G allele) increasing the methylation level by 0.039–0.051 in different cohorts. The positive association between rs3826795 and methylation at CpG site 46,801,642 was also found in the current study, with each risk allele increasing its methylation by 0.029 in obese children, and 0.022 in non-obese children.
Several previous studies showed that HIF3A methylation was associated with BMI. Through the EWAS study of methylation patterns in peripheral blood DNA in one discovery cohort and two replication cohorts, Dick et al. [13] uncovered a specific association between increased BMI and higher methylation levels of 3 CpG sites at the first intron of HIF3A. Demerath et al. [29] validated the results through another EWAS study, and found that HIF3A methylation was associated with 30-year period BMI change in African American adults. Pan et al. [19] found that higher methylation levels at HIF3A CpGs were associated with greater infant weight and adiposity, and further tested an interaction between birth weight and rs3826795 with HIF3A methylation as outcome. Neither study [13, 19] found significant associations between HIF3A SNP and BMI or birth weight, which is in consistency with the current study. In the current study, by testing the nearby SNP and conducting interaction analysis and mediation analysis, we identified that the HIF3A rs3826795 polymorphism interacts with obesity on ALT, and HIF3A DNA methylation could act as a mediator in the effect of rs3826795 on ALT in obese children. This finding provided evidence on the function of HIF3A gene.
The mechanism by which the HIF3A gene could lead to elevation of plasma ALT in obese children is unknown. ALT is clinically used to reflect the defect of liver function, which is related to hepatotoxic fatty acids increase caused by the visceral adipose deposition [30]. ALT elevation is shown to be strongly associated with central adiposity and related features including dyslipidemia, diabetes and hypertension [31]. In a review focusing on obesity and inflammation, Johnson et al. [32] described that in the status of obesity, a variety of cell populations began to exhibit an increased adipose mass and adipocyte diameter, which could lead to cellular hypoxia and further result in pro-inflammatory cytokine production. Rausch et al. [33] found that obesity in male C57BL/6 J mice was associated with increased expression of HIF, and suggested that hypoxia could be a potential contributor to the local and generalized inflammatory state. Chronic low-grade inflammation from visceral adipose tissue is considered as one of the two of the most critical factors (another factor is excess free fatty acids) contributing to liver injury progression. Thus, a possible speculation could be that obesity started a cellular hypoxia status through expanded adipocyte size, and inflammatory cytokines are increased by hypoxia, resulting in the defect of liver function and elevated ALT.
Several limitations of the current study should be noticed. First of all, though we have tested the relation among obesity, ALT, HIF3A SNP and methylation by conducting the interaction analysis and the mediation analysis, the case-control design of the current study means that we cannot assess the causality. However, the identified association provided evidence for further studies. Secondly, known to be a tissue specific biomarker, the HIF3A methylation levels were examined in the current study using peripheral blood samples instead of adipose issues or hepatic issues. As reported by Pfeiffer et al., HIF3A mRNA expression was higher in subcutaneous adipose tissue as compared with visceral adipose tissue, and correlated with parameters of adipose tissue dysfunction [34]. However, several other studies also used peripheral blood to test DNA methylation [12, 29], and supported the use of whole-blood DNA methylation profiling for identification of relevant epigenetic changes. Thirdly, gene-environment interaction analysis and mediation analysis usually need a large sample size. Although only one polymorphism was tested in the current study, multiple tests in the interaction analysis and the mediation analysis have been performed, therefore the result should be interpreted with caution. Besides, for the SNP data, there was a higher proportion of boys in the obese group than in the non-obese group. Although we adjusted for gender in the regression model, the different proportion of male subjects may still influence the result of association. Finally, we did not test cytokines or gene expression data other than DNA methylation. Further studies were needed for understanding the mechanism of the relation among obesity, ALT and the HIF3A gene, and also the mechanism of HIF3A meQTL.
Conclusion
In conclusion, we found that obesity interacts with the HIF3A rs3826795 polymorphism on plasma ALT, and the effect of rs3826795 on ALT in obese children could be mediated by HIF3A DNA methylation. The results provided new evidence to the function of the HIF3A gene and the mechanism linking obesity and ALT. The study would be helpful for future risk assessment and personalized medicine of liver diseases such as NAFLD.
Acknowledgements
The authors thank all the children and their parents for their participation.
Funding
The study was supported by Grants from National Natural Science Foundation of China (81172683) and the Major State Basic Research and Development Program of China (973 Program) (2012CB517501). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Data supporting the findings can be found in the additional supporting file “Supplementary Dataset”.
Abbreviations
- ALT
Alanine aminotransferase
- CpG
cytosine-phosphate-guanine dinucleotides
- EWAS
Epigenome-Wide Association Study
- HIF3A
Hypoxia Inducible Factor 3 Alpha Subunit
- meQTLs
methylation QTLs
- NAFLD
Non-alcoholic fatty liver disease
- QTLs
Quantitative trait loci
- SNP
Single nucleotide polymorphism
Additional file
Dataset of HIF3A polymorphism, methylation, obesity and ALT. In this dataset, each row represents a case and each column represents a variable. This dataset includes 6 variables: (1) log converted plasma ALT; (2) rs3826795 genotype (‘0’ = ‘AA’, ‘1’ = ‘AG’, ‘2’ = ‘GG’); (3) obesity status (‘1’ = ‘non-obese’, ‘2’ = ‘obese’); (4) age_range (years); (5) DNA methylation at 46801699. (XLS 49 kb)
Authors’ contributions
HW and JM conceived and designed the study; SW, JS, YY and YZ collected the laboratory data; SW analyzed the data; SW, YY and NC interpreted the findings; SW drafted the article; NC, JM and HW revised the article for the intellectual content. All authors have read and approved the manuscript.
Ethics approval and consent to participate
All participants and their parents provided their written informed consent. The study was approved by the Ethic Committee of Peking University Health Science Center.
Consent for publication
Not applicable.
Competing interests
The authors all declare that they have no competing financial interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-017-0437-0) contains supplementary material, which is available to authorized users.
Contributor Information
Shuo Wang, Email: wangshuo_20080512@126.com.
Jieyun Song, Email: songjieyun1983@126.com.
Yide Yang, Email: yangyide2007@126.com.
Yining Zhang, Email: zyn1221medical@163.com.
Nitesh V. Chawla, Email: nchawla@nd.edu
Jun Ma, Email: majunt@bjmu.edu.cn.
Haijun Wang, Email: whjun1@bjmu.edu.cn.
References
- 1.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2014;384:766–781. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaskaran K, Douglas I, Forbes H, dos-Santos-Silva I, Leon DA, Smeeth L. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5·24 million UK adults. Lancet. 2014;384:755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.AlKhater SA. Paediatric non-alcoholic fatty liver disease: an overview. Obes Rev. 2015;16:393–405. doi: 10.1111/obr.12271. [DOI] [PubMed] [Google Scholar]
- 4.Anderson EL, Howe LD, Jones HE, Higgins JP, Lawlor DA, Fraser A. The prevalence of non-alcoholic fatty liver disease in children and adolescents: a systematic review and meta-analysis. PLoS One. 2015;10:e0140908. doi: 10.1371/journal.pone.0140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papatheodoridis GV, Goulis J, Christodoulou D, Manolakopoulos S, Raptopoulou M, Andrioti E, et al. High prevalence of elevated liver enzymes in blood donors: associations with male gender and central adiposity. Eur J Gastroenterol Hepatol. 2007;19:281–287. doi: 10.1097/MEG.0b013e328011438b. [DOI] [PubMed] [Google Scholar]
- 6.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in east Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23:5492–5504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marzuillo P, Miraglia del Giudice E, Santoro N. Pediatric fatty liver disease: role of ethnicity and genetics. World J Gastroenterol. 2014;20:7347–7355. doi: 10.3748/wjg.v20.i23.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity: differential variability and differential methylation. Epigenetics. 2013;8:522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego-Durán R, Romero-Gómez M. Epigenetic mechanisms in non-alcoholic fatty liver disease: an emerging field. World J Hepatol. 2015;7:2497–2502. doi: 10.4254/wjh.v7.i24.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith AK, Kilaru V, Kocak M, Almli LM, Mercer KB, Ressler KJ, et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voisin S, Almén MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7:103. doi: 10.1186/s13073-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Song J, Yang Y, Zhang Y, Wang H, Ma J. HIF3A DNA Methylation is associated with childhood obesity and ALT. PLoS One. 2015;10:e0145944. doi: 10.1371/journal.pone.0145944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HJ, Zhang H, Zhang SW, Pan YP, Ma J. Association of the common genetic variant upstream of INSIG2 gene with obesity related phenotypes in Chinese children and adolescents. Biomed Environ Sci. 2008;21:528–536. doi: 10.1016/S0895-3988(09)60013-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Ma J, Zhang S, Hinney A, Hebebrand J, Wang Y, et al. Association of the MC4R V103I polymorphism with obesity: a Chinese case-control study and meta-analysis in 55,195 individuals. Obesity (Silver Spring) 2010;18:573–579. doi: 10.1038/oby.2009.268. [DOI] [PubMed] [Google Scholar]
- 17.Ji CY, Working Group on Obesity in China Report on childhood obesity in China (1)--body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed Environ Sci. 2005;18:390–400. [PubMed] [Google Scholar]
- 18.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan H, Lin X, Wu Y, Chen L, Teh AL, Soh SE, et al. HIF3A association with adiposity: the story begins before birth. Epigenomics. 2015;7:937–950. doi: 10.2217/epi.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu FH, Song JY, Shang XR, Meng XR, Ma J, Wang HJ. The gene-gene interaction of INSIG-SCAP-SREBP pathway on the risk of obesity in Chinese children. Biomed Res Int. 2014;2014:538564. doi: 10.1155/2014/538564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musunuru K, Lettre G, Young T, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3(3):2369–2377. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernaez R, McLean J, Lazo M, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and nutrition examination survey. Clin Gastroenterol Hepatol. 2013;11(9):1183–1190.e2. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037/0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 24.Zou H, Chen Y, Fang W, et al. The mediation effect of health literacy between subjective social status and depressive symptoms in patients with heart failure. J Psychosom Res. 2016;91:33–39. doi: 10.1016/j.jpsychores.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heikkilä M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3avariants in the hypoxia response. Cell Mol Life Sci. 2011;68:3885–3901. doi: 10.1007/s00018-011-0679-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasanen A, Heikkilä M, Rautavuoma K, Hirsilä M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42:1189–1200. doi: 10.1016/j.biocel.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Huang T, Zheng Y, Qi Q, Xu M, Ley SH, Li Y, et al. DNA methylation variants at HIF3A locus, B vitamins intake, and long-term weight change: gene-diet interactions in two US cohorts. Diabetes. 2015;64:3146–3154. doi: 10.2337/db15-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, et al. Epigenome-wide association study (EWAS) of BMI, BMI change, and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24:4464–4479. doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falck-Ytter Y, Younossi ZM, Marchesini G, McCullough AJ. Clinical features and natural history of nonalcoholic steatosis syndromes. Semin Liver Dis. 2001;21:17–26. doi: 10.1055/s-2001-12926. [DOI] [PubMed] [Google Scholar]
- 31.Clark JM, Brancati FL, Diehl AM. The prevalence and etiology of elevated aminotransferase levels in the United States. Am J Gastroenterol. 2003;98:960–967. doi: 10.1111/j.1572-0241.2003.07486.x. [DOI] [PubMed] [Google Scholar]
- 32.Johnson AR, Milner JJ, Makowski L. The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev. 2012;249:218–238. doi: 10.1111/j.1600-065X.2012.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rausch ME, Weisberg S, Vardhana P, Tortoriello DV. Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int J Obes. 2008;32:451–463. doi: 10.1038/sj.ijo.0803744. [DOI] [PubMed] [Google Scholar]
- 34.Susanne P, Jacqueline K, Anna M, et al. Hypoxia-inducible factor 3Agene expression and methylation in adipose tissue is related to adipose tissue dysfunction. Sci Rep. 2016;6:27969. doi: 10.1038/srep27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings can be found in the additional supporting file “Supplementary Dataset”.


