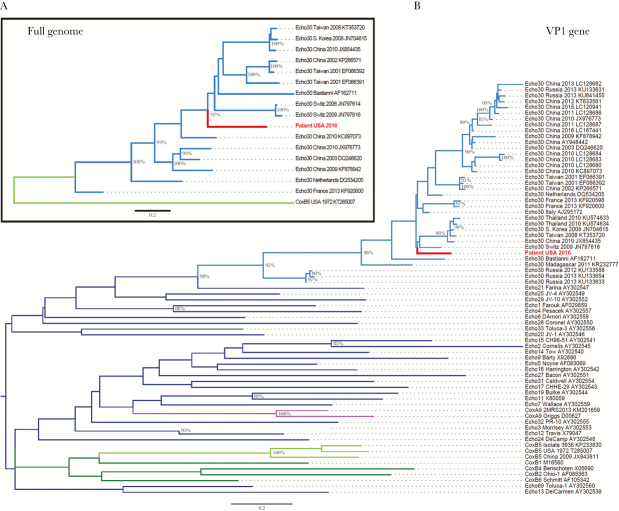
Figure 1.
Maximum likelihood phylogenetic trees of (A) full-length enterovirus genomes and (B) the VP1 gene. (A) The full-length genome sequence identified in this study (“Patient USA 2016”, labeled in red) clusters with sequences belonging to the echovirus 30 subgroup (labeled in light blue). For reference, 1 coxsackie B5 sequence is included (labeled in light green). (B) The VP1 sequence identified in this study (Patient USA 2016, labeled in red) clusters with echovirus 30 sequences (labeled in light blue). Reference VP1 sequences are included from 35 subtypes representing human enterovirus B species: coxsackie B5 (labeled in light green), other coxsackie B subgroups (labeled in dark green), coxsackie A9 (labeled in purple), and other echovirus subgroups (labeled in dark blue). In both trees, branches are named with the enterovirus subtype and location and date of sequence acquisition (clinical samples) or name of reference strain (viral isolates), as well as GenBank accession number. Trees were constructed with 1000 bootstrap replicates, and nodes with at least 80% support are labeled with percent bootstrap support.