Abstract
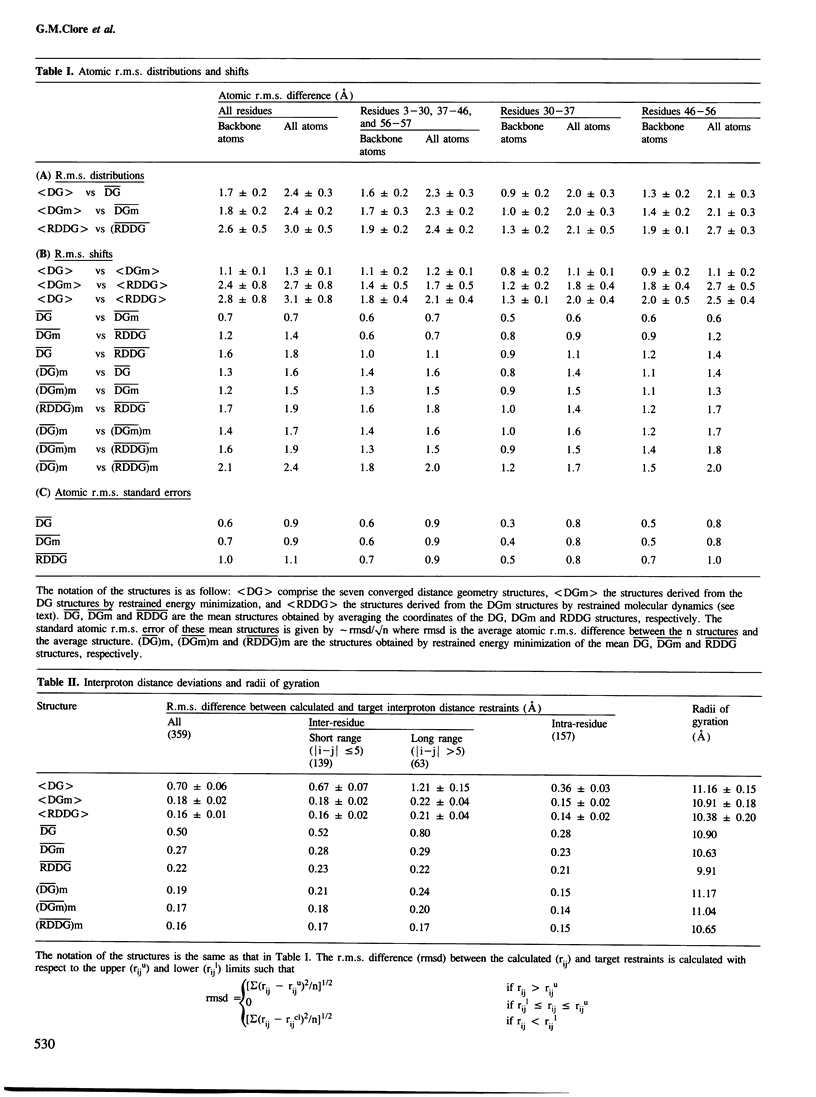
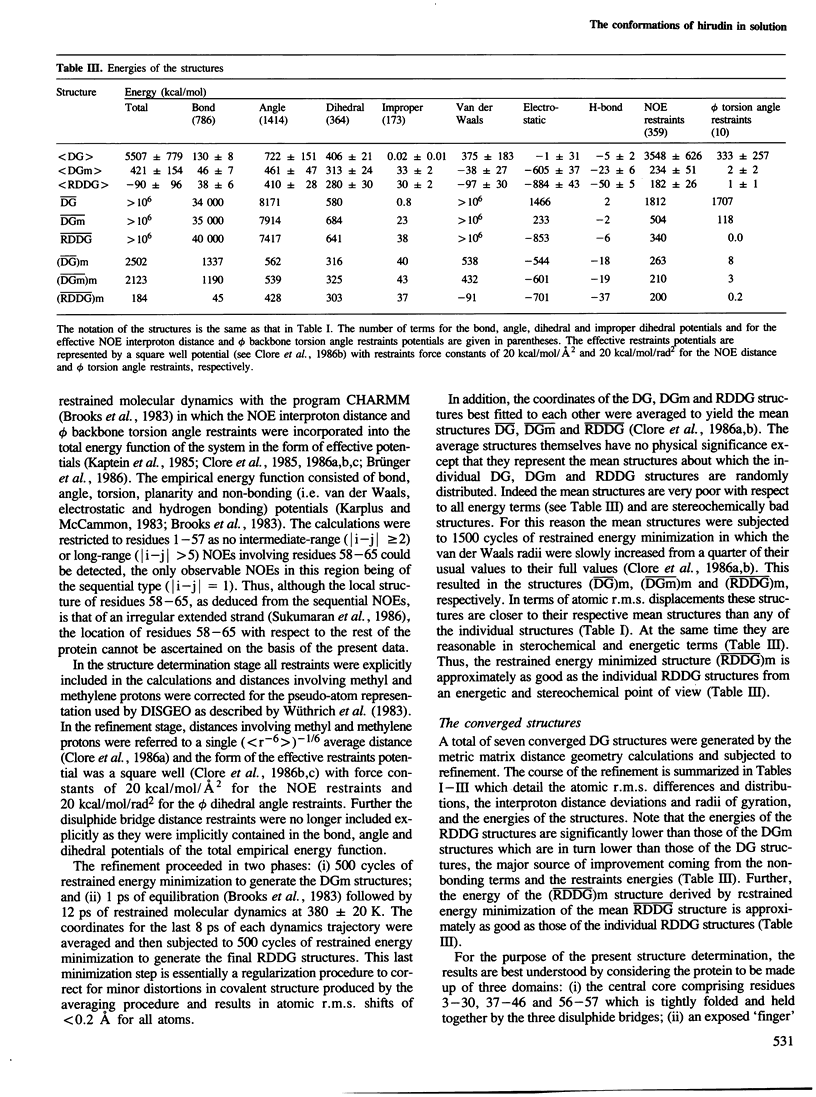
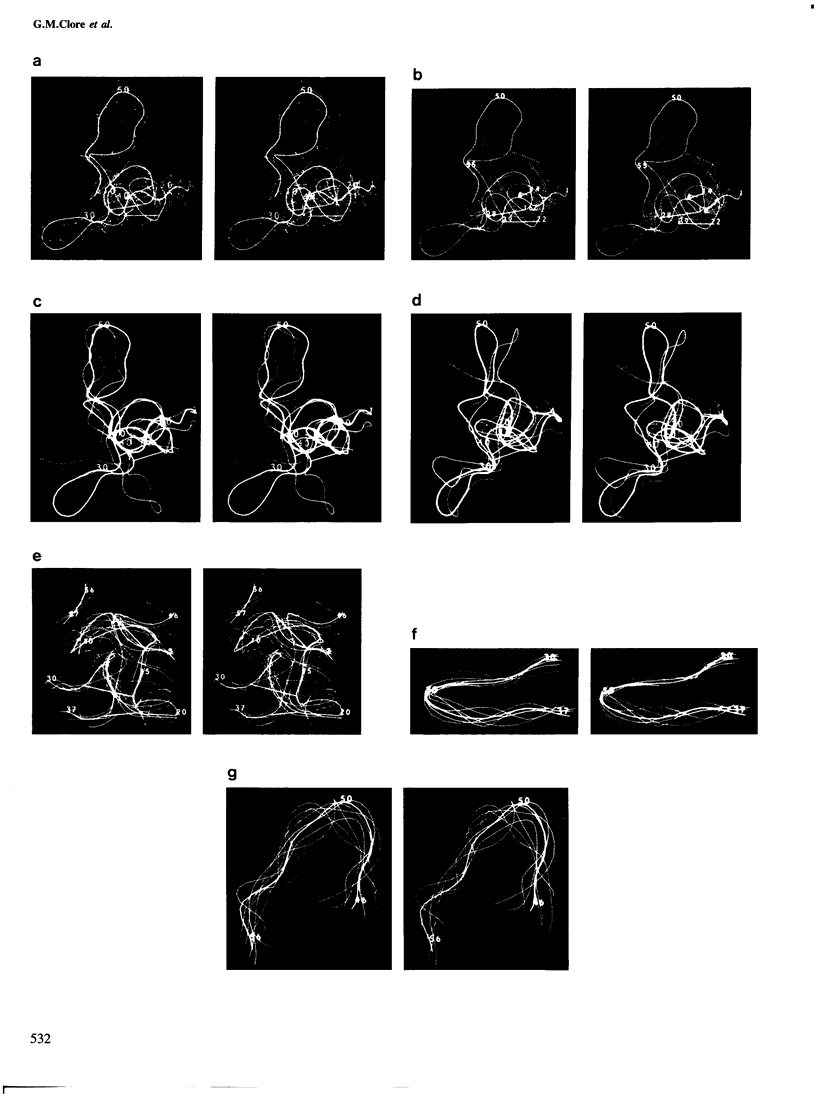
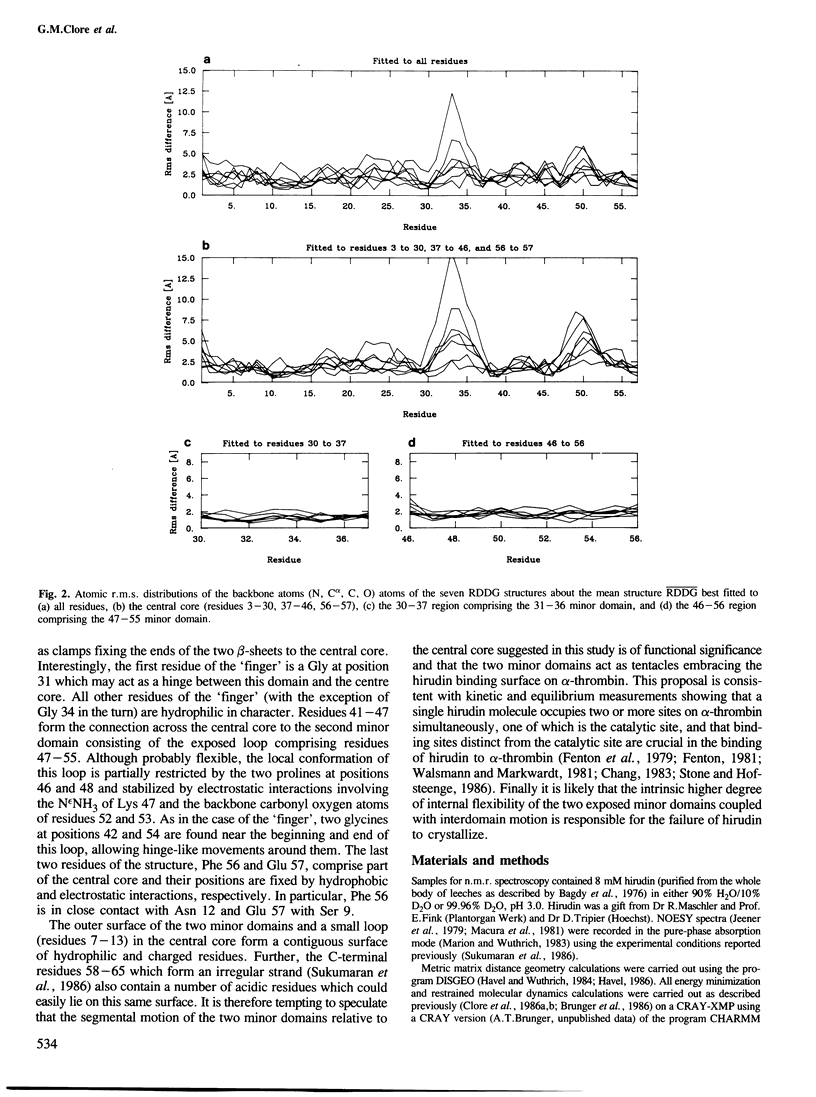
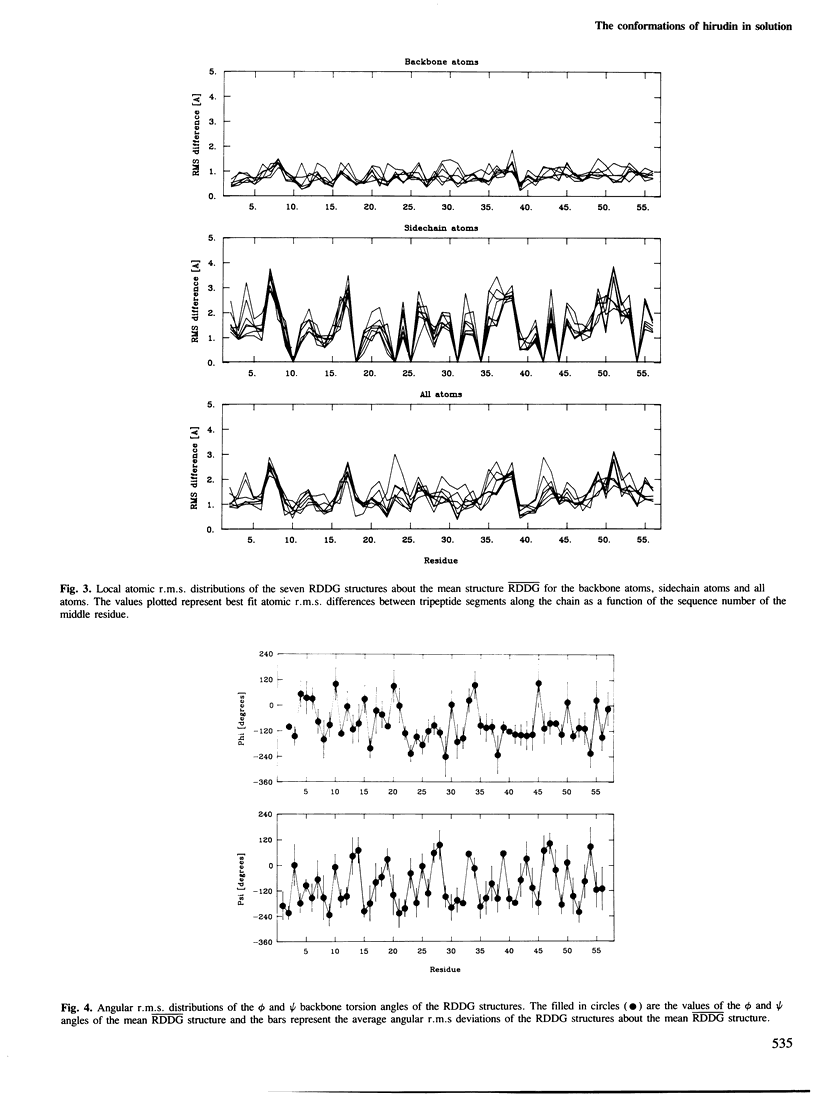
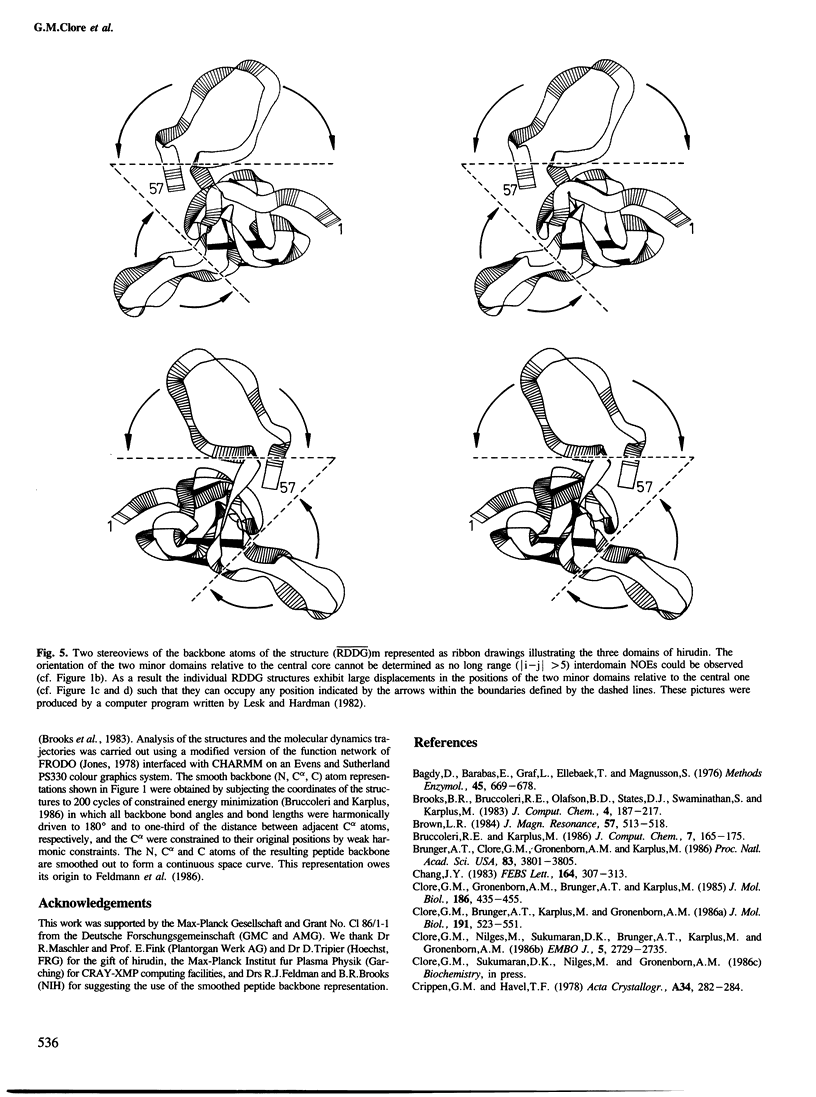
The solution conformations of the protein hirudin have been investigated by the combined use of distance geometry and restrained molecular dynamics calculations. The basis for the structure determination comprised 359 approximate interproton distance restraints and 10 φ backbone torsion angle restraints derived from n.m.r. measurements. It is shown that hirudin is composed of three domains: a central core made up of residues 3-30, 37-46 and 56-57; a protruding `finger' (residues 31-36) consisting of the tip of an antiparallel β sheet, and an exposed loop (residues 47-55). The structure of each individual domain is relatively well defined with average backbone atomic r.m.s. differences of <2 Å between the final seven converged restrained dynamic structures and the mean structure obtained by averaging their coordinates. The orientation of the two minor domains relative to the central core, however, could not be determined as no long-range (ǀi-jǀ>5) interdomain proton–proton contacts could be observed in the two-dimensional nuclear Overhauser enhancement spectra. From the restrained molecular dynamics calculations it appears that the two minor domains exhibit large rigid-body motions relative to the central core.
Keywords: hirudin, solution conformations, nuclear Overhauser effect, interproton distances, distance geometry, restrained molecular dynamics
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdy D., Barabas E., Gráf L., Petersen T. E., Magnusson S. Hirudin. Methods Enzymol. 1976;45:669–678. doi: 10.1016/s0076-6879(76)45057-7. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Clore G. M., Gronenborn A. M., Karplus M. Three-dimensional structure of proteins determined by molecular dynamics with interproton distance restraints: application to crambin. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3801–3805. doi: 10.1073/pnas.83.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. Y. The functional domain of hirudin, a thrombin-specific inhibitor. FEBS Lett. 1983 Dec 12;164(2):307–313. doi: 10.1016/0014-5793(83)80307-x. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Brünger A. T., Karplus M., Gronenborn A. M. Application of molecular dynamics with interproton distance restraints to three-dimensional protein structure determination. A model study of crambin. J Mol Biol. 1986 Oct 5;191(3):523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Solution conformation of a heptadecapeptide comprising the DNA binding helix F of the cyclic AMP receptor protein of Escherichia coli. Combined use of 1H nuclear magnetic resonance and restrained molecular dynamics. J Mol Biol. 1985 Nov 20;186(2):435–455. doi: 10.1016/0022-2836(85)90116-0. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Nilges M., Sukumaran D. K., Brünger A. T., Karplus M., Gronenborn A. M. The three-dimensional structure of alpha1-purothionin in solution: combined use of nuclear magnetic resonance, distance geometry and restrained molecular dynamics. EMBO J. 1986 Oct;5(10):2729–2735. doi: 10.1002/j.1460-2075.1986.tb04557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt J., Seemüller U., Maschler R., Fritz H. The complete covalent structure of hirudin. Localization of the disulfide bonds. Biol Chem Hoppe Seyler. 1985 Apr;366(4):379–385. doi: 10.1515/bchm3.1985.366.1.379. [DOI] [PubMed] [Google Scholar]
- Fenton J. W., 2nd Thrombin specificity. Ann N Y Acad Sci. 1981;370:468–495. doi: 10.1111/j.1749-6632.1981.tb29757.x. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Degryse E., Stefani L., Schamber F., Cazenave J. P., Courtney M., Tolstoshev P., Lecocq J. P. Cloning and expression of a cDNA coding for the anticoagulant hirudin from the bloodsucking leech, Hirudo medicinalis. Proc Natl Acad Sci U S A. 1986 Feb;83(4):1084–1088. doi: 10.1073/pnas.83.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havel T. F., Wüthrich K. An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol. 1985 Mar 20;182(2):281–294. doi: 10.1016/0022-2836(85)90346-8. [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Hafter R., Seemüller U., Gokel J. M., Graeff H. The effect of hirudin on endotoxin induced disseminated intravascular coagulation (DIC). Thromb Res. 1980 Aug 1;19(3):351–358. doi: 10.1016/0049-3848(80)90263-7. [DOI] [PubMed] [Google Scholar]
- Kaptein R., Zuiderweg E. R., Scheek R. M., Boelens R., van Gunsteren W. F. A protein structure from nuclear magnetic resonance data. lac repressor headpiece. J Mol Biol. 1985 Mar 5;182(1):179–182. doi: 10.1016/0022-2836(85)90036-1. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Kline A. D., Braun W., Wüthrich K. Studies by 1H nuclear magnetic resonance and distance geometry of the solution conformation of the alpha-amylase inhibitor tendamistat. J Mol Biol. 1986 May 20;189(2):377–382. doi: 10.1016/0022-2836(86)90519-x. [DOI] [PubMed] [Google Scholar]
- Lent C. New medical and scientific uses of the leech. Nature. 1986 Oct 9;323(6088):494–494. doi: 10.1038/323494a0. [DOI] [PubMed] [Google Scholar]
- Lesk A. M., Hardman K. D. Computer-generated schematic diagrams of protein structures. Science. 1982 Apr 30;216(4545):539–540. doi: 10.1126/science.7071602. [DOI] [PubMed] [Google Scholar]
- Marion D., Wüthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem Biophys Res Commun. 1983 Jun 29;113(3):967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- Markwardt F., Hauptmann J., Nowak G., Klessen C., Walsmann P. Pharmacological studies on the antithrombotic action of hirudin in experimental animals. Thromb Haemost. 1982 Jun 28;47(3):226–229. [PubMed] [Google Scholar]
- Markwardt F. Pharmacology of hirudin: one hundred years after the first report of the anticoagulant agent in medicinal leeches. Biomed Biochim Acta. 1985;44(7-8):1007–1013. [PubMed] [Google Scholar]
- Nilsson L., Clore G. M., Gronenborn A. M., Brünger A. T., Karplus M. Structure refinement of oligonucleotides by molecular dynamics with nuclear Overhauser effect interproton distance restraints: application to 5' d(C-G-T-A-C-G)2. J Mol Biol. 1986 Apr 5;188(3):455–475. doi: 10.1016/0022-2836(86)90168-3. [DOI] [PubMed] [Google Scholar]
- Nowak G., Markwardt F. Influence of hirudin on endotoxin-induced disseminated intravascular coagulation (DIC) in weaned pigs. Exp Pathol (Jena) 1980;18(7-8):438–443. doi: 10.1016/s0014-4908(80)80045-7. [DOI] [PubMed] [Google Scholar]
- Pardi A., Billeter M., Wüthrich K. Calibration of the angular dependence of the amide proton-C alpha proton coupling constants, 3JHN alpha, in a globular protein. Use of 3JHN alpha for identification of helical secondary structure. J Mol Biol. 1984 Dec 15;180(3):741–751. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem Biophys Res Commun. 1983 Dec 16;117(2):479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- Stone S. R., Hofsteenge J. Kinetics of the inhibition of thrombin by hirudin. Biochemistry. 1986 Aug 12;25(16):4622–4628. doi: 10.1021/bi00364a025. [DOI] [PubMed] [Google Scholar]
- Walsmann P., Markwardt F. Biochemische und pharmakologische Aspekte des Thrombininhibitors Hirudin. Pharmazie. 1981 Oct;36(10):653–660. [PubMed] [Google Scholar]
- Williamson M. P., Havel T. F., Wüthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985 Mar 20;182(2):295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Billeter M., Braun W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J Mol Biol. 1983 Oct 5;169(4):949–961. doi: 10.1016/s0022-2836(83)80144-2. [DOI] [PubMed] [Google Scholar]



