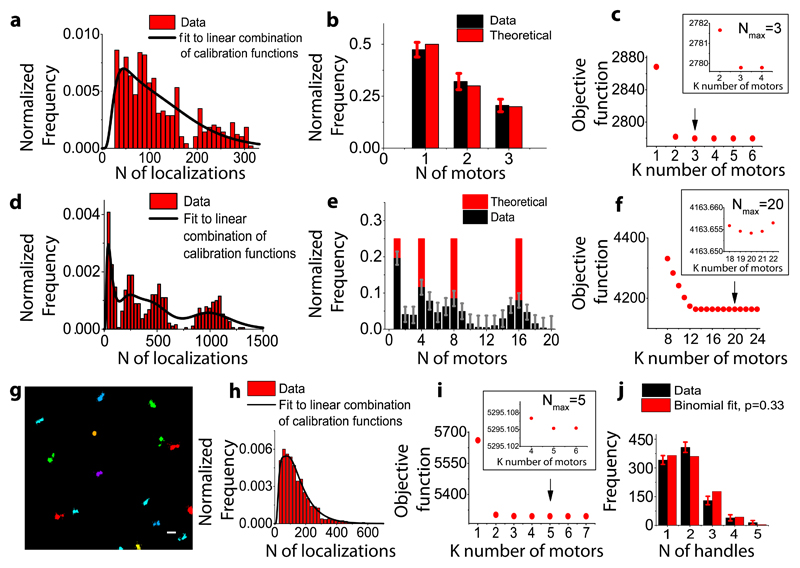
Figure 2. Validation of stoichiometry determination.
(a) Estimation of the stoichiometry for a synthetic sample with known percentage of single, double and triple motors generated from the DNA-origami images, fit to a linear combination of lognormal distributions up to 3 dimers (b) and the objective function for different stoichiometries with a minimum corresponding to 3 motors (Nmax=3) (c) (d) Estimation of the stoichiometry for a synthetic sample with equal percentage (25%) of 1, 4, 8 and 16 motors (2, 8, 16 and 32 GFPs) generated from the DNA-origami images, fit to a linear combination of lognormal distributions up to 20 dimers (e) and the objective function for a number of stoichiometries with a minimum corresponding to 20 motors (Nmax=20) (f). (g) Clustering analysis of STORM images for DNA chassis functionalized with 5 motors. Scale bar 200nm. (h) Distribution showing the total number of localizations per 5 dynein motors (red) and the corresponding fit to a linear combination of log normal functions up to 5 dimers (black line) (i) the objective function for a number of stoichiometries with a minimum corresponding to a stoichiometry of 5 motors (Nmax=5) and (j) The percentage of 1,2,3,4 and 5 motors obtained from the fit in (black) (37% single, 44% two, 14% three, 4% four, 1% five dynein motors) matches to a binomial distribution with a labeling efficiency of p=0.33 (red) (N= 4 independent experiments, number of DNA origami structures analyzed N=934, Reduced ChiSquared=0.0009). Errors bars in (b), (e) and (j) refer to the lower bound to the standard errors based on the Fisher Information Matrix.