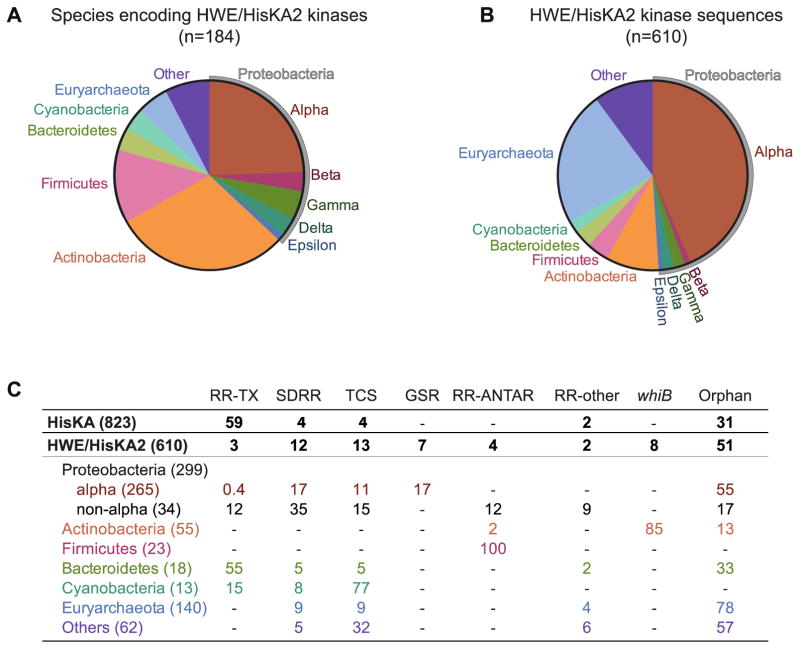
Figure 2. Phylogenetic distribution and genomic context of HWE/HisKA2 histidine kinases.
(A) Distribution of species encoding at least one HWE/HisKA2 kinase, grouped by phylum. (B) Distribution of HWE/HisKA2 kinase sequences, grouped by phylum. Pie charts are comprised of sequences containing either HisKA2 or HWE_HK domains in the Microbial Signal Transduction (MiST) Database version 2.2 (mistdb.com) [5]. Given that these domain models are highly similar, queries with either domain returned sets of sequences that overlapped by ≈70%. The distributions in panels A and B reflect the first 610 sequences in the Mist2.2 database that match either or both domain models. Supplemental File 1 lists the species included in this analysis. (C) Genomic neighborhoods of HWE/HisKA2 kinase genes. We identified the genes adjacent to each HWE/HisKA2 kinase gene tallied in panels A and B. The number in parentheses is the number of kinase genes identified in each phylogenetic group. Numbers in the table are the percentage of sequences identified in each genomic context (e.g. 100% of 23 tallied Firmicute HWE/HisKA2 kinase genes are adjacent to an RR-ANTAR gene). RR-TX: traditional response regulator with receiver domain and a DNA-binding transcriptional regulatory ‘output’ domain; SDRR: single domain response regulator; TCS: two-component locus encoding at least one other histidine kinase; RR-ANTAR: response regulator fused to a RNA-binding antiterminator domain[55,56]; RR-other: receiver domain fused to other ‘output’ domains such as a diguanylate cyclase, phosphodiesterase, or uncharacterized domains; Orphan: no adjacent genes with two-component receiver or kinase domains; whiB: a subset of orphan kinases adjacent to whiB-family transcriptional regulators. In this analysis, hybrid kinases (with a C-terminal receiver domain) were counted with together with non-hybrid kinases. For comparison, we conducted the same analysis on 823 HisKA kinases. Of the classical non-hybrid HisKA kinase genes, 75% are adjacent to a RR-TX gene.