FIGURE 2.
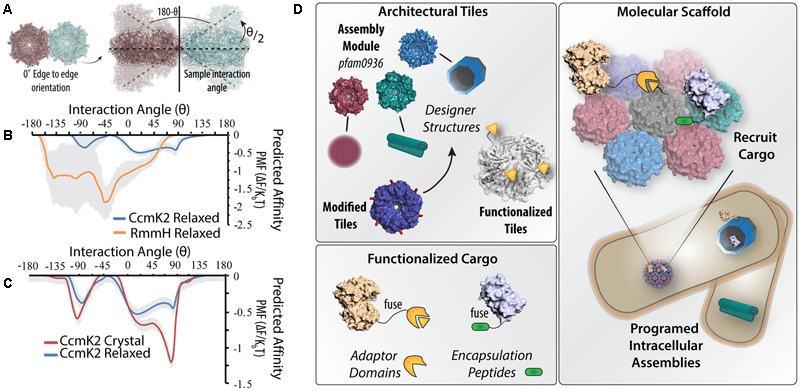
Could molecular-level simulations contribute toward predictive assembly of diverse BMC-H scaffolds? (A) Illustration of design of molecular dynamics simulations where the potential of mean force (PMF) is calculated from two adjacent hexamers. Keeping the relative orientation fixed, the hexamers are systematically rotated out of the plane by an angle 𝜃/2 and the change in PMF is recalculated. (B) Differences in the predicted PMF depending on the inter-hexamer angle are shown for solvated crystal structures of RmmH and CcmK2 (standard deviation between simulations is depicted in gray). (C) Differences in the angular PMF profile can solely arise by comparing the crystal structure versus solvated structure. (D) Illustration of pipeline for constructing BMC-H based programmable nanostructures. BMC-H proteins with different assembly characteristics can be selected from existing homologs (magenta, green, and blue) or created by modification of key residues (red) and modified to encode protein interaction domains (orange). Enzymes and other cargo can be directed to BMC-H assemblies by fusing corresponding ligand domains, or the use of native encapsulation peptides (green). In this manner, it is feasible to envision a diversity of subcellular protein architectures that can be functionalized to scaffold many distinct metabolic or signaling pathways.
