FIGURE 9.
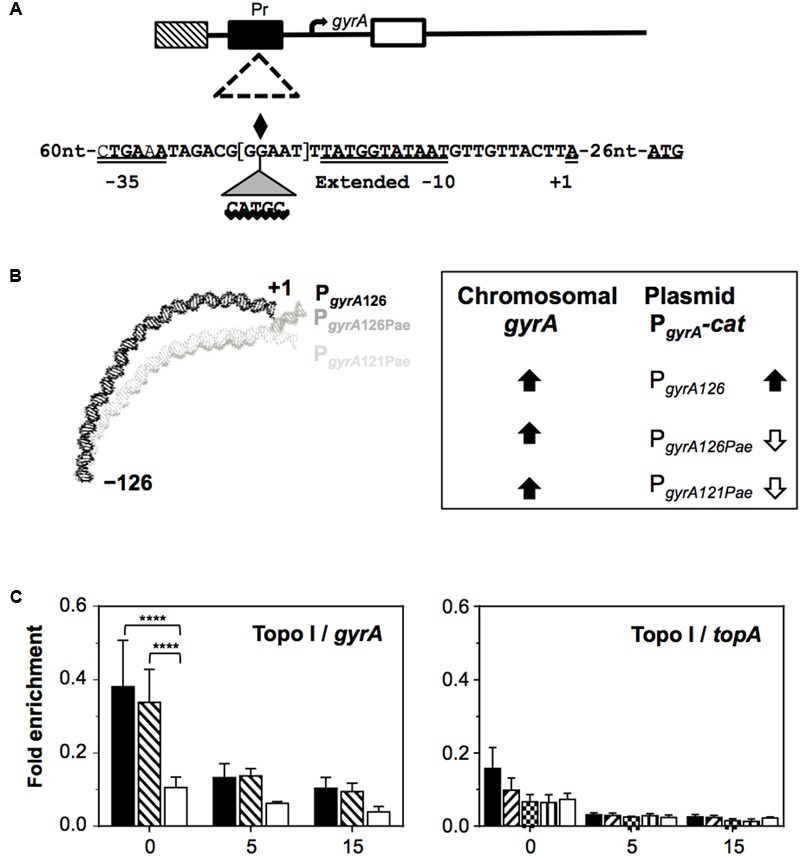
Control of the transcription of DNA topoisomerase genes by supercoiling. (A) Representation of the gyrA coding region and of the regions tested in chromatin immunoprecipitation, showing the sequences of the wild-type PgyrA126, PgyrA126Pae, and PgyrA121Pae derivatives. The –35 and extended –10 boxes, the nucleotide at which transcription is initiated (+1), the center of the intrinsic DNA curvature (diamond), and the location of the inserted CATGC sequence that creates a PaeI restriction site, are all indicated. The five nucleotides deleted in PgyrA121Pae are in brackets. (B) The relaxation-induced up-regulation of PgyrA depends on intrinsic bending: curvature prediction and results obtained from qRT-PCR analysis. (C) Recruitment of Topo I to topA and gyrA upstream sequences. Exponentially growing cells were subjected to chromatin immunoprecipitation using anti-Topo I antibodies; the pulled-down DNA was subsequently analyzed by qPCR. The graphs show the pulldown efficiency (ChIP-DNA/input DNA) for each primer pair. Values are the average ± SD of three independent replicates. ∗∗∗∗P < 0.0001. Taken from Ferrándiz et al. (2014, 2016a), with modifications.
