Abstract
Bone morphogenetic proteins (BMPs) are secreted growth factors that promote differentiation processes in embryogenesis and tissue development. Regulation of BMP signaling involves binding to a variety of extracellular proteins, among which are many von Willebrand factor C (vWC) domain-containing proteins. Although the crystal structure of the complex of crossveinless-2 (CV-2) vWC1 and BMP-2 previously revealed one mode of the vWC/BMP-binding mechanism, other vWC domains may bind to BMP differently. Here, using X-ray crystallography, we present for the first time structures of the vWC domains of two proteins thought to interact with BMP-2: collagen IIA and matricellular protein CCN3. We found that these two vWC domains share a similar N-terminal fold that differs greatly from that in CV-2 vWC, which comprises its BMP-2-binding site. We analyzed the ability of these vWC domains to directly bind to BMP-2 and detected an interaction only between the collagen IIa vWC and BMP-2. Guided by the collagen IIa vWC domain crystal structure and conservation of surface residues among orthologous domains, we mapped the BMP-binding epitope on the subdomain 1 of the vWC domain. This binding site is different from that previously observed in the complex between CV-2 vWC and BMP-2, revealing an alternative mode of interaction between vWC domains and BMPs.
Keywords: bone morphogenetic protein (BMP), collagen, crystal structure, extracellular matrix protein, protein-protein interaction, signaling, CCN proteins, vWC domain
Introduction
Bone morphogenetic proteins (BMPs) 5 are members of the TGF-β superfamily, a group of extracellular growth factors that play important roles in embryonic patterning, differentiation, and homeostasis of various tissue types (1). Many biological processes rely on the normal functioning of BMPs, including the development and maintenance of cartilage and bones, wound repair, and tissue homeostasis, whereas perturbations of BMP signaling are implicated in diseased states such as fibrosis, vascular diseases, and cancer progression (2–8). Secreted dimeric BMPs signal these processes through binding of two type I and two type II serine/threonine kinase receptors, which in turn phosphorylate Smad 1/5/8 proteins and mitogen-activated protein kinases to ultimately regulate gene expression (9). Furthermore, the regulation of BMP signaling involves direct binding by a range of extracellular proteins, including Follistatin, Gremlins, and Noggin, which inhibit the receptor-mediated activity of BMPs (10–15).
Many regulatory proteins, such as the Chordin family members Crossveinless-2 (CV-2), Chordin, and Chordin-like 2 (CHL2), bind BMPs through von Willebrand factor C (vWC) domains. The vWC domain, originally identified in the von Willebrand factor (16), typically contains 75–100 residues, with 10 conserved cysteines. Two cysteine-containing motifs, CXXCXC and CCXXC, lie toward the middle and the C-terminal parts of the domain, respectively (17). X-ray crystallographic and NMR structures have revealed that vWC domains comprise an N-terminal region and two subdomains of SD1 and SD2 linked together by a disulfide bond (17, 18). CV-2, Chordin, and CHL2 each contain multiple vWC domains that interact with BMPs with variable affinities (18–22). Moreover, the various vWC domains have been shown to interact with either or both type I and II receptor-binding sites on BMP-2, thereby resulting in different mechanisms of inhibition of BMP signaling (18, 21, 22).
The crystal structure of the first vWC domain of CV-2 (CV-2 vWC1) in complex with BMP-2 revealed one modality of the vWC/BMP-2 interaction (18). The CV-2 vWC1 binds to BMP-2 through two separate sites of interactions. First, there is high-affinity binding between its N-terminal clip and the type I receptor-binding site on BMP-2 via hydrogen bonds at a nanomolar affinity compatible with the type I receptor/BMP-2 interaction. As shown by its NMR structure in solution at unbound state, this BMP-2-compatible conformation of the N-terminal clip is preformed in the CV-2 vWC domain instead of being induced by complexation (23), suggesting that the N-terminal structure of a vWC domain could be indicative of its binding mechanism with BMPs. Second, there are hydrophobic interactions between its SD1 and the type II receptor-binding site on BMP-2 at a similarly low affinity. The SD2 of CV-2 vWC1 was shown to make no contact with BMP-2 (18). For other BMP-binding vWC domains in Chordin and CHL2, the BMP2-binding epitopes have also been pinned down in the SD1, although structural information of their N termini have hitherto been unavailable to compare their binding modes with that of CV-2 (22).
Two other known BMP-2-binding proteins, Collagen IIA (Col2a) and CCN3, contain largely uncharacterized vWC domains (24–26). Col2a is an alternatively spliced form of procollagen II, expressed in mesenchymal and epithelial cells before differentiation (26, 27). The alternatively spliced exon encodes for a vWC domain, inserted in its non-helical N-terminal propeptide (28). This vWC domain is known to bind BMP-2 and confers anti-BMP-2 activity to Col2a in bioassays (19, 24). The solution NMR structure of Col2a vWC has shown similarities to the fibronectin type 1 domain and CV-2 vWC1 (17, 18, 29). The N-terminal part of Col2a vWC is clearly different from the corresponding part of the CV-2 vWC1, suggesting that Col2a vWC binds BMP-2 through an alternative, as yet uncharacterized mechanism (18).
CCN3 (also known as nephroblastoma overexpressed (Nov)), is a member of the CCN family of matricellular proteins (30). CCN3 has been recognized as a pro-angiogenic factor, a proliferation antagonist, an inhibitor of the bone and cartilage development, and a negative regulator in fibrosis as well as being involved in various types of cancer (31–40). CCN proteins are characterized by their conserved primary structure, comprised of four clearly distinguishable disulfide-rich domains (41). The vWC domain is the second domain in the N-terminal half for CCN proteins after the insulin-like growth factor binding (IB) domain. These two domains are separated by a hinge region from the C-terminal half that contains thrombospondin module 1 (TSP-1) and C-terminal cysteine-knot (CTCK) domains (41). Although it has been demonstrated that CCN3 binds to BMP-2 (25) and that it promotes the mitogen-activated protein kinase signaling, possibly through BMP (42), the exact site of interaction in CCN3 is yet to be identified.
To understand the molecular mechanism of BMP-2 binding by Col2a and CCN3, we have determined crystal structures of vWC domains from both of these proteins and analyzed their interactions with BMP-2. We observe both common and unique features in the crystal structures of these two vWC domains when compared with the existing structure of CV-2 vWC1. Our biochemical analyses demonstrate that Col2a weakly binds to BMP-2 through a novel hydrophobic interaction on the opposite side of its SD1 as compared with CV-2 vWC1. However, despite its overall structural similarities to Col2a vWC, CCN3 vWC does not bind BMP-2.
Results
Crystal structures of Col2a vWC and CCN3 vWC
To begin to make comparisons among vWC domains, we determined the structures of the vWC domains from both Col2a and CCN3 by X-ray crystallography.
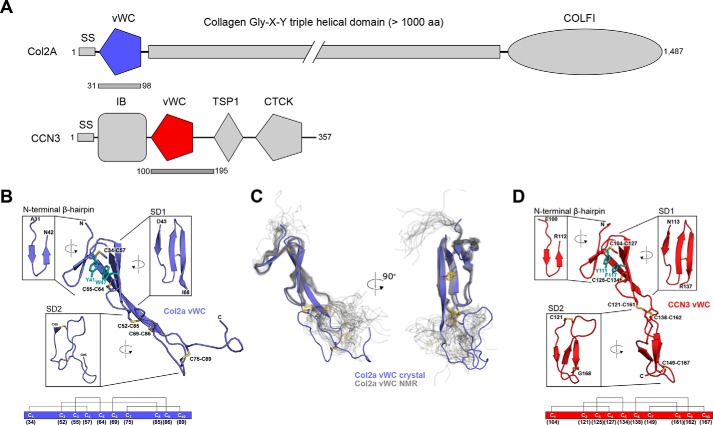
The crystal structure of Col2a vWC was determined through sulfur single wavelength anomalous dispersion (SAD) phasing at a resolution of 1.74 Å (Table 1). As with other vWC domains, the domain has an elongated shape with N and C termini at the opposite ends. As expected, the conserved cysteines are all involved in intramolecular disulfide bonds, with the same connectivity as seen before: Cys1-Cys4, Cys2-Cys8, Cys3-Cys5, Cys6-Cys9, Cys7-Cys10 (superscripted numbers refer to the sequential position of the cysteines in the vWC sequence; Fig. 1). The structure can be subdivided into three parts: an N-terminal β-hairpin region, a subdomain 1 (SD1), and a subdomain 2 (SD2; Fig. 1A). The N-terminal region comprises a two-stranded antiparallel β-sheet that resembles the fold of a β-hairpin, whereas SD1 consists of a three-stranded antiparallel β-sheet with a disulfide (Cys3-Cys5) between the last two strands. The angle between the N-terminal β-hairpin and SD1 is stabilized by a π-stacking interaction formed by two aromatic residues, Tyr-41 and Trp-47 as well as a disulfide bond (Cys1-Cys4) between the first strand of the β-hairpin and the second strand of SD1. SD2 is devoid of regular secondary structure, although it is tethered by one disulfide bond within itself (Cys7-Cys10) and by two disulfides to SD1 (Cys2-Cys8 and Cys6-Cys9). The two molecules in the asymmetric unit align well for most of the structure, with an overall r.m.s.d. of 0.464 Å for 49 of 67 residues; the main differences are found in the first part of SD2 (amino acids (aa) 71–77, r.m.s.d. of 1.05 Å) and in the flexible loop at the C termini (aa 89–96, r.m.s.d. of 2.49 Å) (supplemental Fig. S1A).
Table 1.
X-ray data collection and refinement statistics
Values in parentheses are for the highest resolution shell. Datasets labeled n/a were used for S-SAD and Se-SAD phasing of the corresponding structures.
| Col2a vWC | Col2a vWC | CCN3 vWC | CCN3 vWC V105M/R128M | |
|---|---|---|---|---|
| PDB code | 5NIR | n/a | 5NB8 | n/a |
| Data collection | ||||
| Temperature (K) | 100 | 100 | 100 | 100 |
| Beamline | DLS I24 | DLS I02 | DLS I04–1 | ESRF BM14 |
| Wavelength (Å) | 0.9686 | 1.9000 | 0.9173 | 0.9793 |
| Space group | P212121 | P212121 | P1 | C2 |
| Cell dimensions | ||||
| a, b, c (Å) | 31.9, 60.2, 86.4 | 30.1, 59.6, 86.6 | 38.4, 43.1, 72.3 | 141.7, 43.4, 38.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 75.3, 2.0, 89.9 | 90.0, 98.0, 90.0 |
| Resolution (Å) | 49.37-1.74 | 59.58-1.84 | 28.58-2.10 | 19.01-2.43 |
| (1.75-1.74) | (1.85-1.84) | (2.20-2.10) | (2.59-2.43) | |
| Rmerge | 0.042 (0.709) | 0.053 (0.560) | 0.083 (0.596) | 0.068 (0.269) |
| 〈I〉/σ〈I〉 | 22.7 (2.2) | 24.8 (2.2) | 10.94 (2.60) | 12.04 (4.35) |
| Number of reflections | 112750 | 144119 | 130125 | 65573 |
| CC½a | 1.00 (0.898) | 0.999 (0.886) | 0.998 (0.864) | 0.997 (0.936) |
| Unique reflections | 17667 | 12829 | 25475 | 16452 |
| Multiplicity | 6.4 (5.9) | 11.2 (6.7) | 5.1 (4.7) | 4.0 (4.0) |
| Completeness (%) | 100.0 (100.0) | 90.8 (37.2) | 98.1 (95.3) | 97.2 (96.3) |
| Refinement | ||||
| Resolution (Å) | 49.37-1.74 (1.84-1.74) | 28.58-2.10 (2.18-2.10) | ||
| No. reflections/free | 17616 / 894 | 25446 / 1298 | ||
| Rwork/Rfree | 0.210 / 0.231 | 0.189 / 0.216 | ||
| (0.222 / 0.257) | (0.253 / 0.302) | |||
| No. atoms | ||||
| Protein | 1010 | 2149 | ||
| Ligand/ion | 207 | 31 | ||
| Water | 16 | 155 | ||
| B-factors(Å2) | ||||
| Protein | 35.3 | 49.8 | ||
| Ligand/ion | 48.8 | 92.7 | ||
| Water | 47.2 | 50.5 | ||
| R.m.s.d. | ||||
| Bond lengths (Å) | 0.01 | 0.007 | ||
| Bond angles (°) | 1.18 | 0.872 | ||
a CC½, Pearson correlation coefficient between random half-datasets.
Figure 1.
The tripartite structure of the vWC domains of Col2a and CCN3. A, domain diagrams of Col2a and CCN3 with the two vWC domains colored in purple and red, respectively. The bars underneath the diagrams indicate the parts of the proteins used in this study. IB, insulin-like growth factor binding; TSP-1, thrombospondin module 1; CTCK, C-terminal cysteine-knot. B, ribbon diagram of the crystal structure of Col2a vWC, with insets showing 90° rotated detailed views of each of the three major structural features with start and end residues labeled. Disulfide bonds are labeled and shown in yellow, and key π-π aromatic residues are labeled and shown in teal. The disulfide connectivity is shown schematically at the bottom of the panel with the residue numbers of each cysteine underneath the diagram. C, comparison of the novel crystal structure with the existing NMR ensemble of Col2a vWC (PDB entry 1U5M, thin gray lines) aligned based on the structures of the β-hairpin and SD1. D, ribbon diagram of the CCN3 vWC with insets showing 90° rotated views of each of the three major structural features with the start and end residues labeled. Disulfide bonds are labeled and shown in yellow, and key π-π aromatic residues are labeled and shown in teal. The disulfide connectivity is shown schematically at the bottom of the panel as in B.
The structure of the Col2a vWC domain has previously been solved by NMR (17), and an alignment using only the β-hairpin and SD1 of our crystal structure to the NMR ensemble is shown in Fig. 1B. The β-hairpin and SD1 agree very well between the different NMR models and the crystal structure, with r.m.s.d. of 0.9–1.3 Å. However, we see much poorer overall alignment (r.m.s.d. ∼3.5 Å) when comparing the entire structures or SD2 (supplemental Fig. S2). The disorder in SD2 observed in the NMR ensemble, particularly in the C-terminal tail after the last cysteine, suggests that our crystal structure captured one of many conformations possible for SD2 and that there is flexibility in both SD2 and in its relative orientation to SD1. Additionally, we observe a more extended third β-strand in SD1, a consequence of an extended β-sheet that is formed between the two molecules in the asymmetric unit of the crystal (supplemental Fig. S1B).
Meanwhile, the structure of CCN3 vWC was solved using selenomethionine-labeled vWC, with mutations V105M/R128M to introduce methionines that are otherwise lacking in the domain. This experimentally derived mutant structure was then used to solve the structure of the wild-type vWC domain and that was refined to 2.1 Å resolution (Table 1). Similar to Col2a vWC, the CCN3 vWC comprises an N-terminal β-hairpin, an SD1 of three β-strands, and an SD2 of three β-strands (Fig. 1C), stabilized by five conserved disulfide bonds with the same connectivity. The angle between the β-hairpin and SD1 is also stabilized by a π-π interaction between two aromatic side chains, Tyr-111 and Phe-117, and a disulfide bond (Cys1-Cys4). Although SD2 of Col2a vWC is devoid of regular secondary structure, SD2 of CCN3 vWC consists of a three-stranded antiparallel β-sheet. All four molecules in the asymmetric unit for CCN3 vWC are nearly identical, with r.m.s.d. of 0.121–0.338 Å (supplemental Fig. S3A). The structure of the V105M/R128M mutant domain aligns well with the wild-type vWC domain, whereas its packing in the crystal is very different, suggesting that the orientation between the SD1/SD2 subdomains in CCN3 is relatively stable (data not shown).
Conservation analysis of Col2a vWC and CCN3 vWC
As evolutionary conservation can reflect structural and/or functional importance of individual residues in a protein, we performed such analysis for the newly determined Col2a vWC and CCN3 vWC structures. Given the generally low conservation between different vWC domains and possibly different molecular functions this domain has in different proteins, analysis of sequences from a single species was not deemed to be fruitful. Instead, we used a large number of orthologue sequences across different species to make sure we analyzed conservation between functionally identical domains. We extracted the sequences from the Ensembl genome browser to ensure the widest possible coverage of eukaryotes and removed only the sequences with significant stretches of poorly defined segments.
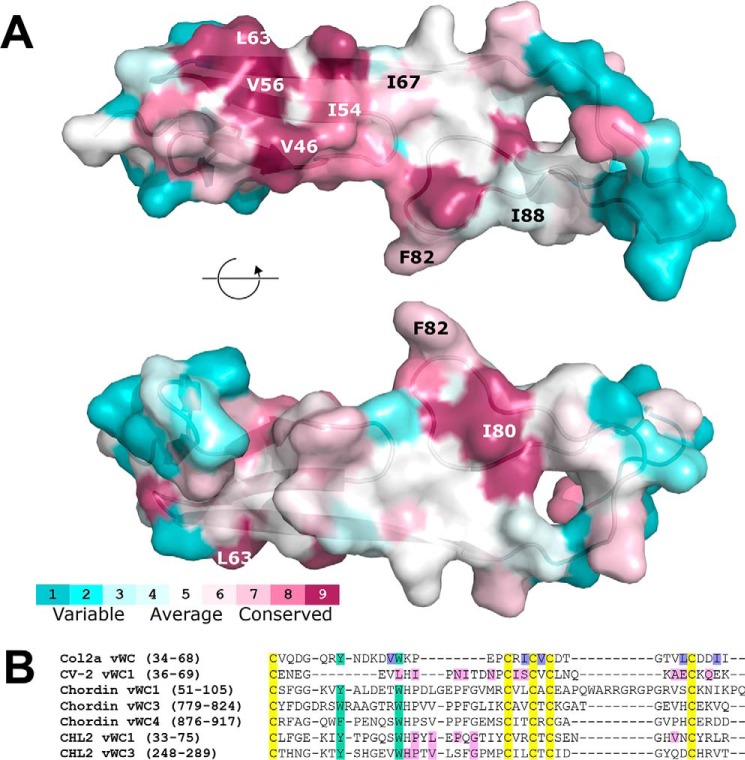
As expected, all 10 cysteines are fully conserved across species in Col2a vWC. The two aromatic residues forming a π-π interaction in SD1, Tyr-41 and Trp-47 in human Col2a vWC, are also conserved for apparent structural importance. The tryptophan residue exists in all species, whereas the phenylalanine residue is found in all but one species (replaced by a histidine in Tetraodon collagen) (supplemental Fig. S7). Besides this structural conservation at the core of the structure, clusters of functional conservation were also observed in various surface positions. These include a hydrophobic patch in the SD1, with two valine residues (Val-46 and Val-56 in human Col2a), a leucine (Leu-63), and an isoleucine (Ile-54), with three substitutions to valines. In SD2 there is another moderately conserved hydrophobic surface consisting of an isoleucine (Ile-80), a phenylalanine (Phe-82), and another isoleucine/valine residue (Ile-88; see Fig. 5A and supplemental Fig. S7).
Figure 5.
Conservation of Col2a vWC structural and binding motifs. A, Conservation of amino acid residues of Col2a among species is shown pictorially using the ConSurf server (consurf.tau.ac.il)6 (65). Residues selected for our mutational analysis are labeled. Full sequence alignments are shown in supplemental Fig. S7. B, sequence alignment of vWC SD1s known to bind BMP-2. Col2a (human, Uniprot P02458), CV-2 (zebrafish, Q5D734), Chordin (mouse, Q9Z0E2), and CHL2 (mouse, Q8VEA6) were aligned using Clustal Omega. Conserved cysteines are highlighted in yellow and conserved π-π aromatic residues are in teal. Residues involved in Col2a binding to BMP-2 identified in this study are highlighted in purple. Residues previously shown to be involved in vWC binding to BMP-2 are highlighted in pink (18, 22).
For CCN3 vWC, the cysteines are also highly conserved across species, with only one exception. In the hedgehog CCN3, three cysteines (Cys1, Cys3, and Cys6) of 10 appear to have mutated. This would have significant consequences for the stability of the domain, as these changes break three of the five disulfides in the domain and would disrupt the structure completely. As in Col2a, both aromatic residues involved in the stabilization of the N-terminal β-hairpin and SD1 are highly conserved in CCN3 vWC. The tyrosine is invariable in all but 5 of 69 sequences, whereas the phenylalanine is even more conserved with substitutions to tyrosine found in just two species. However, unlike Col2a vWC, conservation of solvent-exposed residues on the CCN3 vWC domain is less evident. The only cluster with relatively conserved residues is in the SD2. This site is composed of Pro-145, Lys-153, and Trp-165 in CCN3 vWC (supplemental Fig. S8), but in the absence of detectable binding to BMP-2 we have not studied the possible functional role of these residues.
Structural comparisons among Col2a vWC, CCN3 vWC, and CV-2 vWC1
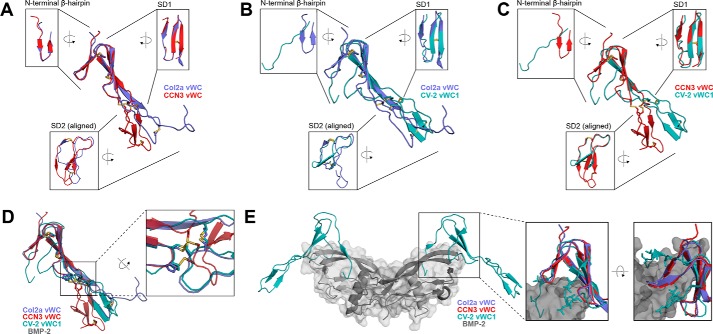
Besides our new structures and the Col2a NMR structure, the only other vWC structure available is that of CV-2 vWC1 bound to BMP-2 (16). Fig. 2, A–C, show pair-wise comparisons of these three structures (A, Col2a/CCN3; B, Col2a/CV-2; C, CCN3/CV-2). Generally, these three structures share common subdomain architectures and disulfide bond connections. Disulfide Cys1-Cys4 is between the N-terminal clip (CV-2) or the first strand of N-terminal β-hairpin (Col2a and CCN3) and the second strand of SD1. Cys3-Cys5 connects the second and third strand in SD1. The disulfide bond of Cys2-Cys8 links SD1 with SD2, whereas Cys6-Cys9 and Cys7-Cys10 form interstrand disulfide bonds at the top and bottom of the SD2, respectively. Although the connectivities of the five disulfide bonds are identical in all three vWC structures, detailed comparisons reveal subtle differences in their positions. In CCN3 vWC, only the Cys1-Cys4 and Cys3-Cys5 in the N-terminal half are in the same positions as in Col2a and CV-2 vWCs. The lack of a proline residue before the Cys2 in CCN3 (Pro-51 in Col2a and Pro-28 in CV-2) has caused the Cys2-Cys8, and subsequently Cys9-Cys6, to shift their positions by one residue (Fig. 2D and supplemental Fig. S4, B and C). This proline residue is only conserved in about a quarter of all human vWC domains, suggesting these proteins will have similar spatial organization of disulfide bonds as in CCN3 vWC (supplemental Fig. S5).
Figure 2.
Structural and BMP-2-binding epitope comparisons among vWC domains. A–C, overlays of the Col2a (purple) and CCN3 (red), Col2a (purple) and CV-2 (turquoise, PDB entry 3BK3), and CCN3 (red) and CV-2 (turquoise) vWCs, respectively, with insets show 90° rotated views of each of the three major structural features. D, overlay of the three vWC domains, with a zoomed-in inset highlighting the disulfide bonds that link SD1 and SD2. E, structure of BMP-2 (gray with semi-transparent surface) in complex with vWC domain from CV-2 (turquoise). The insets show overlays of Col2a vWC SD1 and CCN3 vWC SD1 with CV-2 vWC1 SD1 bound to BMP-2 (gray). Residues in CV-2 involved in BMP-2 binding are shown as sticks.
Although the subdomain boundaries and disulfide connectivities are conserved among the three currently available vWC structures, they display significant differences as well. The angles formed between SD1 and SD2 display great variability among the three structures, varying by up to 50°. This is not particularly surprising, as the NMR ensembles of the structures of Col2a vWC and CV-2 vWC1 domains in solution suggests significant flexibility in the linker between the subdomains. Additionally, the structural changes caused by the proline before the second cysteine (Cys2) and the shift of the two disulfide positions in Col2a and CV-2 vWCs are likely to play a role in the relative orientation between SD1 and SD2. Previous investigations of CV-2 and CHL2 vWC domains have demonstrated that SD1 subdomains are responsible for BMP-2 binding, and SD2 subdomains appear to play no direct role in this (18, 22).
Superimposition of the structures shows that the N-terminal halves of Col2a vWC and CCN3 vWC domains are more similar to each other than they are to the corresponding part of CV-2 vWC1. Two major structural differences suggest that the Col2a and CCN3 vWC domains cannot bind BMP-2 in the same manner as CV-2 vWC1. First, the N-terminal regions of Col2a and CCN3 vWCs have more regular secondary structures compared with that in CV-2 vWC1 (Fig. 2, A–C). The double-stranded antiparallel β-sheet found in both Col2a vWC and in CCN3 vWC resembles a hairpin instead of the clip-like fold in CV-2 vWC1. The angle between the N-terminal β-hairpin and SD1 is stabilized via a disulfide bond Cys1-Cys4 as well as the aromatic interaction between Tyr-41 and Trp-47 in Col2a and Tyr-111 and Phe-117 in CCN3. Although the equivalent disulfide bond is also observed in CV-2 vWC1, the aromatic residues are absent, and the N-terminal part of the domain forms a more loosely packed structure. As shown by Zhang et al. (18) the N-terminal clip of CV-2 vWC1 is very important for the high affinity binding to BMP-2 on the type I receptor-binding site via hydrogen bonds. Even though the hydrogen-bond formation is through the main chain atoms in CV-2 vWC1 and thus does not require sequence conservation, the folded β-hairpin structure in Col2a and CCN3 vWC domains would not allow their N-terminal regions to bind BMP-2 in the same manner as CV-2 vWC1. Second, the first of the three antiparallel strands in SD1 is part of a regular β-sheet in Col2a vWC and CCV3 vWC, whereas in CV-2 vWC1 it forms an extended loop structure that is the other major element of the interface with the hydrophobic type II receptor site of BMP-2. Superimposition of the Col2a vWC SD1 and CCN3 vWC SD1 with CV-2 vWC1 SD1 bound to BMP-2 demonstrates how the absence of the N-terminal clip and the extended loop in the first strand of SD1 prevents the same interaction with BMP-2 (Fig. 2E). Taken together, it is unlikely that Col2a vWC and CCN3 vWC can bind to BMP-2 in the same manner as CV-2 vWC1.
Characterizations of vWC/BMP-2 interactions
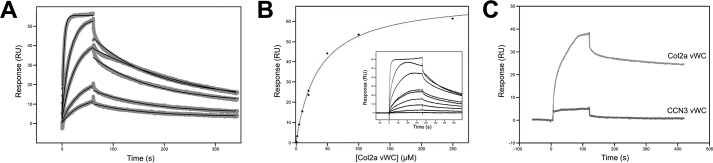
To support our structural analyses that suggest new modes of vWC/BMP-2 interactions, we set about characterizing the binding of Col2a and CCN3 vWC domains to BMP-2 using surface plasmon resonance (SPR). Purified recombinant mature domain of BMP-2 was covalently immobilized on the surface of the sensor chip, and purified Col2a and CCN3 vWC domains were flowed over the sensor chip. As has been shown previously (19, 24), Col2a vWC was found to bind to BMP-2, albeit at low affinity. The binding rate constants koff (7 × 10−3 s−1) and kon (7 × 102 m−1s−1) were determined by fitting kinetic data to a 1:1 binding model, resulting in a KD of 10 μm (KD = koff/kon) (Fig. 3A). This calculated binding affinity was validated by equilibrium SPR experiments, resulting in a KD of 42 μm (Fig. 3B).
Figure 3.
Characterization of the binding of Col2a and CCN3 vWC domains to immobilized BMP-2 by SPR analysis. A, kinetic analysis of Col2a vWC binding to BMP-2. BMP-2 was immobilized, whereas 10–250 μm Col2a vWC was flowed over the sensor chip surface. Raw data are shown in light gray with fitting curves overlaid in black. Each dissociation and association phase was fit separately to give a kon of 700 ± 300 m−1s−1, koff of 7 × 10−3 ± 3 × 10−3 s−1, and KD (koff/kon) of 11 μm (n = 2). B, equilibrium binding of Col2a vWC to BMP-2. Equilibrium response was plotted as dose dependence of Col2a vWC and fit to give a KD of 42 ± 6 μm (n = 2). Raw traces are shown as the inset. C, binding of 40 μm Col2a vWC and CCN3 to BMP-2 are shown as indicated in the graph. RU, resonance units.
However, despite having a similar SD1 structure to Col2a vWC, CCN3 vWC did not show any detectable binding to BMP-2 at a concentration range up to 40 μm when analyzed by SPR in identical conditions used for Col2a vWC (Fig. 3C). Given that we use highly purified and homogenous proteins, we are very confident that the interaction between BMP-2 and CCN3 (and most likely other CCN proteins) is not mediated by the vWC domain, or at most its contribution to the binding is very small.
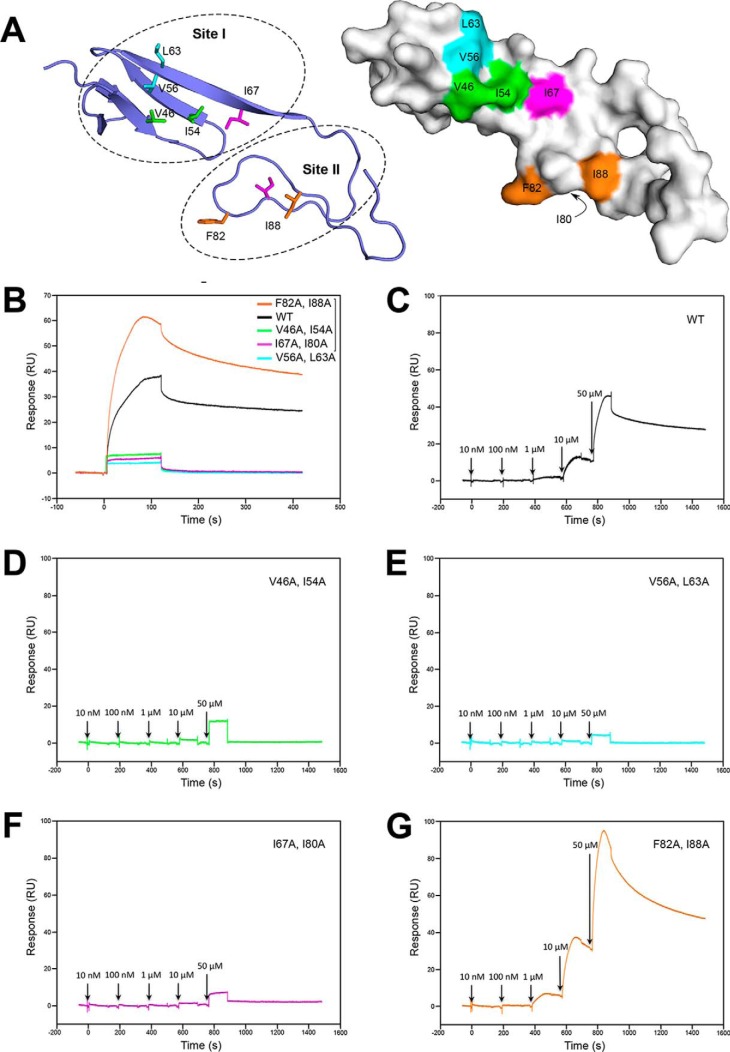
Based on these structural and biophysical data, it is clear that the residues responsible for BMP-2 binding in the Col2a vWC are unlikely to be the same as in CV-2 vWC1. Given that structurally more similar CCN3 vWC is not binding to BMP-2, the BMP-2 binding residues in Col2a would not be conserved in CCN3 either. To define the binding epitope of Col2a vWC to BMP-2, we identified two conserved clusters of surface-exposed hydrophobic residues: I, Val-46, Ile-54, Val-56, Leu-63, and Ile-67 in SD1; II, Ile-80, Phe-82, and Ile-88 in SD2 (Figs. 4A and 5A). Site I in SD1 is in the opposite side to the CV-2 vWC1 epitope, whereas the site in the SD2 is directly below the CV-2 vWC1 epitope. We mutated these residues in pairs (based on closeness in the 3D structure) to alanines, generating four mutants of Col2a vWC (Fig. 4A). All the mutants were expressed and purified as the wild-type domain, appearing to produce well folded proteins.
Figure 4.
Mutational analysis of the BMP-2-binding epitope of Col2a vWC. A, ribbon diagram of the Col2a vWC crystal structure showing surface-exposed hydrophobic residues selected for mutagenesis in site I and II. B–G, binding of Col2a vWC variants to immobilized BMP by SPR analysis. B, SPR sensorgrams of 40 μm wild-type (black) and mutants (colored as in A) of Col2a vWC binding to BMP-2. C–G, SPR sensorgrams of increasing concentrations of Col2a vWC variants (colored as in A and B) ranging from 10 nm to 50 μm sequentially injected at the time points indicated by arrows. RU, response units.
The effect of these mutations on the interaction with BMP-2 was monitored by SPR. These SPR experiments revealed that all three mutants containing mutations in site I, V46A+I54A, V56A+L63A, and I67A+I80A, showed no interaction with BMP-2, whereas the F82A+I88A mutant, with both residues in site II, retained its ability to bind BMP-2 (Fig. 4, B–G). Because the interaction between the Col2a vWC and BMP-2 is weak (KD 10–42 μm), it is not surprising that mutation of one or two residues involved in the binding can be enough to completely disrupt the interaction. Taken together, it can be inferred that the binding epitope of Col2a vWC to BMP-2 resides in site I, on the opposite side of SD1 to the CV-2 vWC1 epitope, with residues Val-46 and/or Ile-54, Val-56, and/or Leu-63, and Ile-67 contributing to the hydrophobic interactions with BMP-2.
Discussion
We have determined the structures of the vWC domains of collagen IIA and CCN3 using X-ray crystallography. Through structural comparison with CV-2 vWC1, we predict that Col2a vWC and CCN3 vWC are not able to bind to BMP-2 in the same manner as CV-2 vWC1. Using SPR, we analyzed the binding of the two vWC domains of collagen IIA and CCN3 to BMP-2. Kinetic rate constants of the Col2a vWC/BMP-2 interaction were measured, and the binding affinity was determined using both kinetic and equilibrium SPR data. CCN3 vWC, on the other hand, showed no binding to BMP-2 at the same concentration range used as Col2a vWC. Leveraging our structural analysis, we identified the binding epitope of Col2a vWC to BMP-2 as a hydrophobic patch on the opposite side of SD1 as the previously characterized BMP-2 binding site in CV-2 vWC1.
Both Col2a and CCN3 vWC domains show a typical tripartite structure of an N-terminal region and two subdomains SD1 and SD2. The non-globular, extended confirmation of the vWC domains potentially allows a great deal of flexibility for the relative orientation of SD2 to SD1, as has been observed in solution NMR structures of Col2a vWC and CV-2 vWC1 (17, 23). Given this flexibility, the hydrogen-bond network between β-sheets in adjacent molecules in the asymmetric units of both Col2a and CCN3 vWCs must be key contributions to the well diffracting crystals (supplemental Figs. S1B and S3B). As commonly observed in most cysteine-rich extracellular proteins, the formation of the disulfide bonds are largely responsible for the stability of this small domain, reflected in the complete conservation of these residues. One of the unique features that we observed in Col2a vWC and CCN3 vWC, but not in CV-2 vWC1, is the π-π interaction that locks the orientation of the N-terminal β-hairpin. The two aromatic residues that form this π-π interaction, one in the N-terminal region and the other in the SD1, are conserved in more than half (162 of 259) of the vWC domains found in human proteins (supplemental Fig. S6), including vWC domains known to bind BMP-2 (Fig. 5B). These vWC domains would potentially share the N-terminal structure of the β-hairpin with Col2a vWC and CCN3 vWC rather than the extended clip-like fold found in CV-2 vWC1.
The two major binding epitopes for BMP-2 in CV-2 vWC1, the N-terminal clip and the extended first strand in SD1, are both absent in our Col2a vWC and CCN3 vWC structures. It was suggested by Zhang et al. (18) that the Col2a vWC could undergo major conformational rearrangements to make the same epitope accessible for binding to BMP-2. However, a solution NMR structure of the CV-2 vWC1 in its unbound state showed that the conformations of both epitopes were predefined, with the angle of the N-terminal clip relative to SD1 being fixed and the extended first strand in SD1 being very rigid (23). Instead, our mutational study on Col2a vWC has uncovered a novel BMP-2 binding epitope on the opposite side of SD1 as compared with that in the CV-2 vWC1. Sequence conservation analysis has also shown that this epitope is highly conserved across different species, with a score of 9 (9 to 1 from conserved to variable) for Val-46, Val-56, and Leu-63, 8 for Ile-54, and 7 for Ile-67 (Fig. 5A and supplemental Fig. S7). Because the structure of the CCN3 vWC revealed no conservation of either of the two CV-2 vWC1 epitopes nor the residues involved in binding in Col2a vWC, it is not surprising that we did not observe any binding of CCN3 vWC to BMP-2. However, the association between the CCN family and the BMPs is clearly evident in the literature (25, 43–48); therefore, this interaction must be mediated through one or more of the three other domains (IB (insulin-like growth factor binding), thrombospondin module 1 (TSP1), or CTCK (C-terminal cysteine-knot)) in the CCN proteins.
This new mechanism of vWC/BMP interaction provides insights into the possible binding modes of other BMP-binding vWC domains (Table 2 and Fig. 5B). For Chordin and CHL2, their vWC1 and vWC3 were both shown to bind BMP-2 in the SD1 (22). Residues involved in the binding in CHL2 vWC1 and vWC3 were also identified by mutational analysis by Fujisawa et al. (22). Sequence alignment with Col2a vWC and CV-2 vWC1 suggests that these residues lie in the first strand of SD1 (Fig. 5B), and given that CHL2 and Chordin possess 7–10 more residues in this region, it is possible that they form an even more extended loop toward the BMP-2 and share the same epitope with CV-2 vWC1. However, the two aromatic residues that form the π-π interaction between the N-terminal region and SD1 may make it conformationally more preferable to access BMP-2 from the opposite side as we have shown for the Col2a vWC. It is also possible that these vWC domains bind BMP-2 through yet another epitope, and further studies are necessary to parse this question.
Table 2.
BMP-2 binding affinities of full-length and/or vWC domains of Col2a, CV-2, Chordin, and CHL2
Besides CV-2, previous studies have revealed inhibition mechanisms of other known antagonists of BMP signaling, typically through blocking the type I and type II receptor-binding sites. Similar to CV-2, both Noggin and Gremlin-2 use their flexible and extended N-terminal regions to reach into the type I receptor-binding site of BMP together with additional interfaces that overlap with the type II receptor site (11, 14). Gremlin-1 also shares the binding sites of both type I and II receptors, although not involving its N termini (12). Despite similar inhibition mechanisms, the stoichiometries of these antagonists with their ligand BMPs are quite diverse. One dimeric Noggin binds to one dimeric BMP with a 1:1 stoichiometry, whereas two molecules of CV-2 vWC1 bind to each protomer of a dimeric BMP with a 2:1 stoichiometry. For Gremlin-1 and -2, distinguished features of elongated fibrils of oligomeric complexes have been suggested where each protomer of a dimeric Gremlin binds to one protomer of BMP, forming an alternating, open-ended oligomer (12, 14). To elucidate the binding mechanism by Col2a, future studies should include mutagenesis of receptor-binding sites on BMP-2, complex crystallization, and small-angle X-ray scattering analysis.
The affinity of Col2a vWC for BMP-2 we obtained is much lower when comparing with the vWC domains in CV-2, Chordin, and CHL2 (Table 2). However, the affinity of full-length Chordin and CHL2, which allows multiple vWC domains to simultaneously bind BMP-2, is 7.5–2500-fold higher than their individual vWC domains (Table 2) (18, 21, 22, 49). In vivo, collagen molecules self-associate into trimeric coils and to further oligomeric fibrils, with the vWC domain containing N-terminal propeptide accessible on the surface of the collagen fibrils (24). Multimeric collagen could allow two vWC domains to interact simultaneously with a dimeric BMP-2, forming oligomeric fibrils like the Gremlins (12, 14) and thereby increase the apparent affinity between the two proteins due to avidity effects. In fact, Col2a vWC expressed as a GST fusion protein has been shown to bind BMP-2 with nanomolar affinity (24). Furthermore, previous studies with Xenopus laevis oocytes demonstrated anti-BMP-2 activity with full-length collagen IIA but not with isolated Col2a vWC domain (19), suggesting that avidity effects play a significant role in the biological efficacy of Col2a vWC. BMPs are also thought to bind to fibrillar collagen directly, and such interactions could exist alongside vWC binding and further stabilize such complex. Furthermore, like all TGF-β family growth factors, BMPs are synthesized as larger precursors, and the propeptides can interact with components of the extracellular matrix (50). Col2a vWC interaction is, therefore, one of the many ways BMPs interact with extracellular matrix.
As collagen is a major structural component of the extracellular matrix, its binding of BMP-2 could affect the localization and generation of morphogenetic gradients during development. Procollagen IIA is expressed in epithelial and mesenchymal cells before differentiation, suggesting it may play an important role in tissue differentiation and body patterning (26–28). In our biological activity assays, we did not observe inhibition or stimulation of the BMP-2 signaling by monomeric Col2a vWC (data not shown). This could be due to the low-affinity binding by a single vWC domain rather than the trimeric full-length collagen. Alternatively, the binding to BMP-2 by collagen could be neutral with regard to BMP-2 bioactivity, with collagen instead serving as a scaffold for localization and storage of BMP-2, assisting in its gradient formation and presenting it to other extracellular regulators. Although we have not been able to characterize the BMP-2–Col2a vWC complex structurally, our data suggest a distinct mode of interaction that has been observed with CV-2 and noggin. Localization of the BMP-2-binding epitope on the relatively flat surface of the Col2a vWC domain suggests that this interaction cannot involve both the type I and II receptor-binding sites, which are located on the opposite sides of the growth factor.
Experimental procedures
Cloning, expression, and purification
Human collagen IIA vWC domain (residues 31–98, Uniprot P02458) and rat CCN3 vWC domain (residues 100–195, Uniprot Q9QZQ5) were expressed in BL21(DE3) Escherichia coli using the pHAT3 and pBAT4 vectors (51), respectively. DNA for the Col2a vWC construct was codon-optimized for E. coli and synthesized by PCR using overlapping oligonucleotides (all oligonucleotide sequences are listed in the supplemental material and Table S1). CCN3 was amplified from cDNA (a kind gift from Dr. Paul Kemp). vWC mutants were generated by a two-step PCR protocol using overlapping oligonucleotide across the mutated regions and cloned into expression vectors as the wild-type proteins. All constructs were confirmed by dideoxy sequencing. For protein expression, E. coli BL21(DE3) competent cells were transformed with the expression plasmid and grown on L-agar plates in the presence of 100 μg/ml ampicillin overnight. The following day cells from the plate were cultured in 2-YT medium with 100 μg/ml ampicillin at 37 °C under agitation. Cells were induced with 400 μm isopropyl-β-d-thiogalactoside at an A600 nm of 0.8 and left to express the proteins for 4 h. Both proteins were expressed solubly despite the presence of 10 cysteines that form disulfide bridges in the folded proteins.
His-tagged Col2a vWC was purified by nickel-nitrilotriacetic acid affinity chromatography followed by cleavage of the His tag by thrombin protease and final purification by reversed phase chromatography (ACE® 5 C8-300). CCN3 vWC was purified by anion exchange (HiTrap™ Q HP, GE Healthcare) and size exclusion chromatographies (HiLoad™ 16/60 Superdex™ 75, GE Healthcare) followed by reversed phase chromatography (ACE® 5 C8-300). Both proteins were lyophilized and resuspended in water after reverse-phase chromatography. Purified proteins were analyzed by MALDI mass spectrometry and were observed to have the mass for proteins in which all cysteine residues have been oxidized to cystines. To obtain experimental phases for CCN3 vWC crystal structure determination, the V105M/R128M mutant protein was labeled with selenomethionine using the metabolic suppression method and purified like the wild-type domain. Production of BMP-2 has been described previously (12).
Crystallization and structure determination
Crystals of the seleno-l-methionine-labeled CCN3 vWC mutant grew in 3.4 m NaCl, 100 mm MES, pH 6.8, in 1 μl + 1 μl drops. Crystals were cryoprotected with 10% glycerol and cryocooled in liquid nitrogen. Data from these crystals were collected at ESRF (European Synchrotron Radiation Facility) beamline BM14 at selenium peak at a wavelength of 0.97963 Å. Data were processed using XDS, and the experimental phases were derived from the SAD data using autoSHARP (52). The initial model was built automatically using ARP/wARP (53). Crystals of unlabeled CCN3 vWC domain grew in 0.8 m potassium/sodium tartrate, 0.4 m NaCl, 0.1 m imidazole, pH 8.0. The crystals were cryo-protected in 30% glycerol and cryo-cooled in liquid nitrogen. X-ray diffraction data were collected at Diamond Light Source, beamline i04-1 using a Pilatus 2M detector (Dectris, Baden-Daettwil, Switzerland) at wavelength 0.9173 Å. X-ray diffraction data of CCN3 vWC were indexed, integrated, and scaled using XDS (54). Two datasets obtained from separate crystals were merged using XSCALE (54). Molecular replacement was performed by AMoRe (55) using the experimentally phased CCN3 vWC V105M/R128M mutant structure as the search model. Refinement was performed using Refmac 5.5 (56) and Phenix 1.8.4 (57). Coot (58) was used for iterative model building and initial validation.
Col2a vWC domain was crystallized by mixing 1 μl of protein at a concentration of 10 mg/ml and 1 μl of reservoir solution (0.1 m HEPES, pH 7.5, 40% PEG 200) in a sitting-drop-vapor-diffusion setup. Experimental phases were obtained from sulfur SAD. Anomalous data were collected at a wavelength of 1.9 Å at beamline I04 of Diamond Light Source and processed via autoPROC (59). Phases were obtained using ShelxC, ShelxD, and ShelxE (60) through the HKL2MAP (61) interface. An initial model was obtained through automated model building using Buccaneer (62). Further iterative model building was performed using autoBUSTER (63) and Coot (58). This preliminary model was then used in molecular replacement (PHASER; Ref. 64) to extend the resolution to data collected at a wavelength of 0.9686 Å at beamline I24 at Diamond Light Source and refined as above.
Statistics of X-ray data collection and structure refinement are shown in Table 1. Both Col2a and CCN3 vWC structures have been deposited in the Protein Data Bank with accession codes 5NIR and 5NB8, respectively.
Conservation analysis
The degree of evolutionary conservation of the amino acids in the Col2a vWC and CCN3 vWC domains was analyzed using the ConSurf server (consurf.tau.ac.il/2016) 6 (65), to reveal the regions that are important for the function and/or structure of the protein. Sequences of Col2a vWC and CCN3 vWC across species were extracted from the Ensembl database (release 86, ensembl.org/index.html) (66), aligned, and scored for position-specific conservation by the ConSurf Server and mapped onto the surfaces of the corresponding structures (65).
SPR
SPR binding assays were performed on a BIAcore T100 instrument (GE Healthcare). HBS-EP+ buffer (20 mm HEPES, 150 mm NaCl, 3 mm EDTA, 0.05% (v/v) Surfactant P20, pH 7.4) with the addition of 1 mg/ml CM-Dextran salt was used as the running buffer at a flow rate of 30 μl/min. BMP-2 (residues 283–396) was directly immobilized on a CM5 sensor chip (GE Healthcare) via amine coupling. Wild type and mutants of Col2a vWC as well as the CCN3 vWC were used as analytes. The surface was regenerated with 4 m guanidinium chloride. The sensograms were corrected for unspecific binding by subtracting the signal from the reference channel. Data were analyzed with BIAcore T100 Evaluation Software 2.0.3 (GE Healthcare) and proFit (QuantumSoft) using kinetic and equilibrium equations for a 1:1 interaction.
Author contributions
M. H. conceived and coordinated the study. E.-R. X., E. E. B., and M. H. designed the experiments. E. E. B. performed the experiments with Col2a vWC domain. E.-R. X. performed the experiments with CCN3 vWC domain and Col2a vWC mutants. G. F. determined the structure of Col2a vWC domain. M. H. determined the original CCN3 vWC domain. All authors analyzed the results, wrote, revised, and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Katharina Ravn for the production of BMP-2 and Dr. May Marsh for help with crystallization. We are grateful for access to and support at the X-ray crystallographic and Biophysics facilities at the Department of Biochemistry. We thank the Diamond Light Source and the beamline staff for access to beamline i02, i04-1 and i24 (proposals mx6886 and mx7141, respectively), and ESRF (European Synchrotron Radiation Facility) and their beamline staff for access to beamline BM14.
This work was supported by Cambridge Overseas Trust and China Scholarship Council through a postgraduate scholarship (to E.-R. X.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S8.
The atomic coordinates and structure factors (codes 5NB8 and 5NIR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- BMP
- bone morphogenetic protein
- vWC
- von Willebrand factor C domain
- Col2a
- collagen type IIα
- CV-2
- Crossveinless-2
- CHL2
- Chordin-like 2
- SPR
- surface plasmon resonance
- SAD
- single wavelength anomalous dispersion
- SD1 and -2
- subdomains 1 and 2
- r.m.s.d.
- root mean square deviation(s).
References
- 1. Hogan B. L. (1996) Bone morphogenetic proteins in development. Curr. Opin. Genet. Dev. 6, 432–438 [DOI] [PubMed] [Google Scholar]
- 2. Reddi A. (2003) Cartilage morphogenetic proteins: role in joint development, homoeostasis, and regeneration. Ann. Rheum. Dis. 62, ii73–ii78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simic P., and Vukicevic S. (2005) Bone morphogenetic proteins in development and homeostasis of kidney. Cytokine Growth Factor Rev. 16, 299–308 [DOI] [PubMed] [Google Scholar]
- 4. Moura J., da Silva L., Cruz M. T., and Carvalho E. (2013) Molecular and cellular mechanisms of bone morphogenetic proteins and activins in the skin: potential benefits for wound healing. Arch. Dermatol. Res. 305, 557–569 [DOI] [PubMed] [Google Scholar]
- 5. Tobin J. F., and Celeste A. J. (2006) Bone morphogenetic proteins and growth differentiation factors as drug targets in cardiovascular and metabolic disease. Drug Discov. Today. 11, 405–411 [DOI] [PubMed] [Google Scholar]
- 6. Manson S. R., Song J. B., Guo Q., Liapis H., and Austin P. F. (2015) Cell type specific changes in BMP-7 expression contribute to the progression of kidney disease in patients with obstructive uropathy. J. Urol. 193, 1860–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cai J., Pardali E., Sánchez-Duffhues G., and ten Dijke P. (2012) BMP signaling in vascular diseases. FEBS Lett. 586, 1993–2002 [DOI] [PubMed] [Google Scholar]
- 8. Bandyopadhyay A., Yadav P. S., and Prashar P. (2013) BMP signaling in development and diseases: a pharmacological perspective. Biochem. Pharmacol. 85, 857–864 [DOI] [PubMed] [Google Scholar]
- 9. Miyazono K., Kamiya Y., and Morikawa M. (2010) Bone morphogenetic protein receptors and signal transduction. J. Biochem. 147, 35–51 [DOI] [PubMed] [Google Scholar]
- 10. Balemans W., and Van Hul W. (2002) Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev. Biol. 250, 231–250 [PubMed] [Google Scholar]
- 11. Groppe J., Greenwald J., Wiater E., Rodriguez-Leon J., Economides A. N., Kwiatkowski W., Affolter M., Vale W. W., Izpisua Belmonte J. C., and Choe S. (2002) Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 420, 636–642 [DOI] [PubMed] [Google Scholar]
- 12. Kišonait M., Wang X., and Hyvönen M. (2016) Structure of Gremlin-1 and analysis of its interaction with BMP-2. Biochem. J. 473, 1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salazar V. S., Gamer L. W., and Rosen V. (2016) BMP signalling in skeletal development, disease, and repair. Nat. Rev. Endocrinol. 12, 203–221 [DOI] [PubMed] [Google Scholar]
- 14. Nolan K., Kattamuri C., Rankin S. A., Read R. J., Zorn A. M., and Thompson T. B. (2016) Structure of Gremlin-2 in complex with GDF5 gives insight into DAN-family-mediated BMP antagonism. Cell Rep. 16, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nolan K., Kattamuri C., Luedeke D. M., Deng X., Jagpal A., Zhang F., Linhardt R. J., Kenny A. P., Zorn A. M., and Thompson T. B. (2013) Structure of protein related to DAN and Cerberus: insights into the mechanism of bone morphogenetic protein antagonism. Structure 21, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mancuso D. J., Tuley E. A., Westfield L. A., Worrall N. K., Shelton-Inloes B. B., Sorace J. M., Alevy Y. G., and Sadler J. E. (1989) Structure of the gene for human von Willebrand factor. J. Biol. Chem. 264, 19514–19527 [PubMed] [Google Scholar]
- 17. O'Leary J. M., Hamilton J. M., Deane C. M., Valeyev N. V., Sandell L. J., and Downing A. K. (2004) Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand factor type C module) from collagen IIA. J. Biol. Chem. 279, 53857–53866 [DOI] [PubMed] [Google Scholar]
- 18. Zhang J. L., Qiu L. Y., Kotzsch A., Weidauer S., Patterson L., Hammerschmidt M., Sebald W., and Mueller T. D. (2008) Crystal structure analysis reveals how the chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev. Cell. 14, 739–750 [DOI] [PubMed] [Google Scholar]
- 19. Larraín J., Bachiller D., Lu B., Agius E., and Piccolo S., and De Robertis E. M. (2000) BMP-binding modules in chordin: a model for signalling regulation in the extracellular space. Development 127, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garcia Abreu J., Coffinier C., Larraín J., Oelgeschläger M., and De Robertis E. M. (2002) Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 287, 39–47 [DOI] [PubMed] [Google Scholar]
- 21. Zhang J.-L., Huang Y., Qiu L.-Y., Nickel J., and Sebald W. (2007) von Willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J. Biol. Chem. 282, 20002–20014 [DOI] [PubMed] [Google Scholar]
- 22. Fujisawa T., Huang Y., Sebald W., and Zhang J. L. (2009) The binding of von Willebrand factor type C domains of Chordin family proteins to BMP-2 and Tsg is mediated by their SD1 subdomain. Biochem. Biophys. Res. Commun. 385, 215–219 [DOI] [PubMed] [Google Scholar]
- 23. Fiebig J. E., Weidauer S. E., Qiu L. Y., Bauer M., Schmieder P., Beerbaum M., Zhang J. L., Oschkinat H., Sebald W., and Mueller T. D. (2013) The clip-segment of the von Willebrand domain 1 of the BMP modulator protein crossveinless 2 is preformed. Molecules 18, 11658–11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Y., Oganesian A., Keene D. R., and Sandell L. J. (1999) Type IIA procollagen containing the cysteine-rich amino propeptide is deposited in the extracellular matrix of prechondrogenic. J. Cell Biol. 144, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Minamizato T., Sakamoto K., Liu T., Kokubo H., Katsube K., Perbal B., Nakamura S., and Yamaguchi A. (2007) CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem. Biophys. Res. Commun. 354, 567–573 [DOI] [PubMed] [Google Scholar]
- 26. Ryan M. C., and Sandell L. J. (1990) Differential expression of a cysteine-rich domain in the amino-terminal propeptide of type II (cartilage) procollagen by alternative splicing of mRNA. J. Biol. Chem. 265, 10334–10339 [PubMed] [Google Scholar]
- 27. Sandell L. J., Nalin A. M., and Reife R. A. (1994) Alternative splice form of type II procollagen mRNA (IIA) is predominant in skeletal precursors and non-cartilaginous tissues during early mouse development. Dev. Dyn. 199, 129–140 [DOI] [PubMed] [Google Scholar]
- 28. Oganesian A., Zhu Y., and Sandell L. J. (1997) Type IIA procollagen amino propeptide is localized in human embryonic tissues. J. Histochem. Cytochem. 45, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 29. Williams M. J., Phan I., Harvey T. S., Rostagno A., Gold L. I., and Campbell I. D. (1994) Solution structure of a pair of fibronectin type 1 modules with fibrin binding activity. J. Mol. Biol. 235, 1302–1311 [DOI] [PubMed] [Google Scholar]
- 30. Leask A., and Abraham D. J. (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J. Cell Sci. 119, 4803–4810 [DOI] [PubMed] [Google Scholar]
- 31. Lin C. G., Leu S. J., Chen N., Tebeau C. M., Lin S. X., Yeung C. Y., and Lau L. F. (2003) CCN3 (NOV) is a novel angiogenic regulator of the CCN protein family. J. Biol. Chem. 278, 24200–24208 [DOI] [PubMed] [Google Scholar]
- 32. Bleau A. M., Planque N., Lazar N., Zambelli D., Ori A., Quan T., Fisher G., Scotlandi K., and Perbal B. (2007) Antiproliferative activity of CCN3: involvement of the C-terminal module and post-translational regulation. J. Cell. Biochem. 101, 1475–1491 [DOI] [PubMed] [Google Scholar]
- 33. Rydziel S., Stadmeyer L., Zanotti S., Durant D., Smerdel-Ramoya A., and Canalis E. (2007) Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J. Biol. Chem. 282, 19762–19772 [DOI] [PubMed] [Google Scholar]
- 34. Janune D., Kubota S., Nishida T., Kawaki H., Perbal B., Iida S., and Takigawa M. (2011) Novel effects of CCN3 that may direct the differentiation of chondrocytes. FEBS Lett. 585, 3033–3040 [DOI] [PubMed] [Google Scholar]
- 35. Abd El Kader T., Kubota S., Janune D., Nishida T., Hattori T., Aoyama E., Perbal B., Kuboki T., and Takigawa M. (2013) Anti-fibrotic effect of CCN3 accompanied by altered gene expression profile of the CCN family. J. Cell Commun. Signal. 7, 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohgawara T., Kubota S., Kawaki H., Kurio N., Abd El Kader T., Hoshijima M., Janune D., Shimo T., Perbal B., Sasaki A., and Takigawa M. (2011) Association of the metastatic phenotype with CCN family members among breast and oral cancer cells. J. Cell Commun. Signal. 5, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cui L., Xie R., Dang S., Zhang Q., Mao S., Chen J., Qu J., and Zhang J. (2014) NOV promoted the growth and migration of pancreatic cancer cells. Tumour Biol. 35, 3195–3201 [DOI] [PubMed] [Google Scholar]
- 38. McCallum L., Price S., Planque N., Perbal B., Pierce A., Whetton A. D., and Irvine A. E. (2006) A novel mechanism for BCR-ABL action: stimulated secretion of CCN3 is involved in growth and differentiation regulation. Blood 108, 1716–1723 [DOI] [PubMed] [Google Scholar]
- 39. Ouellet V., and Siegel P. M. (2012) CCN3 modulates bone turnover and is a novel regulator of skeletal metastasis. J. Cell Commun. Signal. 6, 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang C. C., Lin B. R., Wu T. S., Jeng Y. M., and Kuo M. L. (2014) Input of microenvironmental regulation on colorectal cancer: Role of the CCN family. World J. Gastroenterol. 20, 6826–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bork P. (1993) The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett. 327, 125–130 [DOI] [PubMed] [Google Scholar]
- 42. Kawaki H., Kubota S., Suzuki A., Suzuki M., Kohsaka K., Hoshi K., Fujii T., Lazar N., Ohgawara T., Maeda T., Perbal B., Takano-Yamamoto T., and Takigawa M. (2011) Differential roles of CCN family proteins during osteoblast differentiation: involvement of Smad and MAPK signaling pathways. Bone 49, 975–989 [DOI] [PubMed] [Google Scholar]
- 43. Abreu J. G., Ketpura N. I., Reversade B., and De Robertis E. M. (2002) Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat. Cell Biol. 4, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakamura Y., Weidinger G., Liang J. O., Aquilina-Beck A., Tamai K., Moon R. T., and Warman M. L. (2007) The CCN family member Wisp3, mutant in progressive pseudorheumatoid dysplasia, modulates BMP and Wnt signaling. J. Clin. Invest. 117, 3075–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen T. Q., Roestenberg P., van Nieuwenhoven F. A., Bovenschen N., Li Z., Xu L., Oliver N., Aten J., Joles J. A., Vial C., Brandan E., Lyons K. M., and Goldschmeding R. (2008) CTGF inhibits BMP-7 signaling in diabetic nephropathy. J. Am. Soc. Nephrol. 19, 2098–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maeda A., Nishida T., Aoyama E., Kubota S., Lyons K. M., Kuboki T., and Takigawa M. (2009) CCN family 2/connective tissue growth factor modulates BMP signalling as a signal conductor, which action regulates the proliferation and differentiation of chondrocytes. J. Biochem. 145, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ono M., Inkson C. A., Kilts T. M., and Young M. F. (2011) WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J. Bone Miner. Res. 26, 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pal A., Huang W., Li X., Toy K. A., Nikolovska-Coleska Z., and Kleer C. G. (2012) CCN6 modulates BMP signaling via the Smad-independent TAK1/p38 pathway, acting to suppress metastasis of breast cancer. Cancer Res. 72, 4818–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Troilo H., Zuk A. V., Tunnicliffe R. B., Wohl A. P., Berry R., Collins R. F., Jowitt T. A., Sengle G., and Baldock C. (2014) Nanoscale structure of the BMP antagonist chordin supports cooperative BMP binding. Proc. Natl. Acad. Sci. U.S.A. 111, 13063–13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wohl A. P., Troilo H., Collins R. F., Baldock C., and Sengle G. (2016) Extracellular regulation of bone morphogenetic protein activity by the microfibril component fibrillin-1. J. Biol. Chem. 291, 12732–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peränen J., Rikkonen M., Hyvönen M., and Kääriäinen L. (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 [DOI] [PubMed] [Google Scholar]
- 52. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., and Paciorek W. (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 53. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kabsch W. (2010) XDS. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Cryst. A50, 157–163 [Google Scholar]
- 56. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., and Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 57. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 59. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G. (2011) Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 61. Pape T., and Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 [Google Scholar]
- 62. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 63. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., PW and Roversi P., Sharff A., Smart O. S., Vonrhein C. W. T. O. (2016) BUSTER version 2.11.5. Glob. Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 64. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T., and Ben-Tal N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aken B. L., Ayling S., Barrell D., Clarke L., Curwen V., Fairley S., Fernandez Banet J., Billis K., García Girón C., Hourlier T., Howe K., Kähäri A., Kokocinski F., Martin F. J., Murphy D. N., et al. (2016) The Ensembl gene annotation system. Database (Oxford) pii: baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.