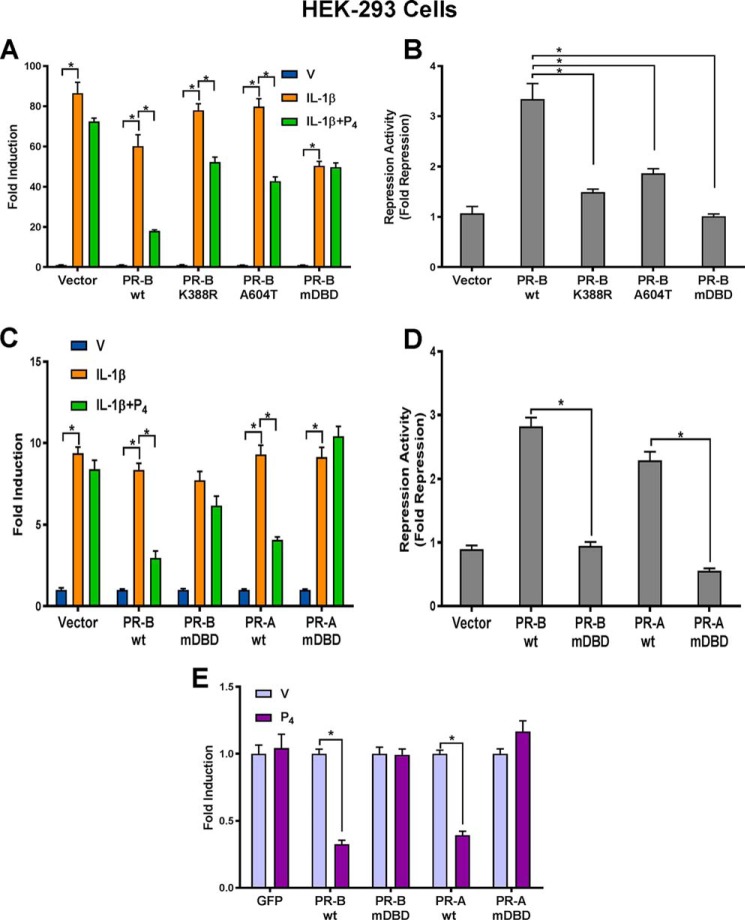
Figure 1.
Progesterone–PR–mediated repression of NF-κB reporter activity in HEK-293 cells is lost by mutagenesis of the PR DBD. A, HEK-293 cells were cotransfected with NF-κB-luciferase reporter constructs, Renilla luciferase plasmid, and expression vectors of PR-BWT and PR-B mutants, including PR-B K388R (sumoylation mutant), PR-B A604T (dimerization mutant), and PR-BmDBD P-box mutant. One day after transfection, cells were treated with DMSO (V), IL-1β (10 ng/ml), or IL-1β plus P4 (100 nm) for 6 h. B, repression activity/-fold repression of data shown in A was calculated by dividing the “-fold induction” for PR-BWT and each PR-B mutant in response to IL-1β by the -fold induction in response to IL-1β + P4. C, to further study the effects of DBD mutations on transrepression activity of PR-B and PR-A, HEK-293 cells were cotransfected with PR-BWT, PR-AWT, or the corresponding DBD mutants and with NF-κB-luciferase reporter constructs and Renilla luciferase plasmid. One day after transfection, cells were treated with DMSO (V), IL-1β (10 ng/ml), or IL-1β plus P4 (100 nm) for 6 h. D, repression activity of PR-BWT and PR-AWT and corresponding DBD mutants shown in C was calculated for cells transfected with each PR isoform and mutant by dividing the -fold induction in response to IL-1β by the -fold induction in response to IL-1β + P4. E, HEK-293 cells were cotransfected with Gal4–p65 C-terminal transactivation domain expression vector, GAL4 luciferase reporter, and PRWT or PRmDBD expression vector, as indicated. One day after transfection, cells were treated with DMSO (V) or P4 (100 nm) for 24 h. Cells from each experiment were then harvested, and firefly luciferase and Renilla luciferase activities were assayed. Relative luciferase activities were calculated by normalizing firefly luciferase activity to Renilla luciferase activity in the same samples to correct for transfection efficiencies. Data for experiments shown in A–E are the mean ± S.E. (error bars) of three replicate determinations for each treatment group. *, significant (p < 0.05) difference between samples.