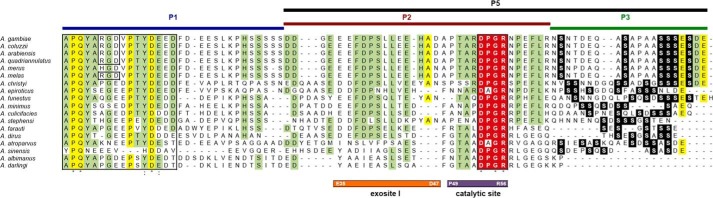
Figure 4.
Multiple alignment of cE5/anophelin family members from 18 Anopheles species. The conserved N-terminal region and the DPGR tetrapeptide (highlighted in red) are boxed. Serine residues in the C-terminal region are shown in white lettering on a black background. Residues conserved in all or at least in 75% of the aligned sequences are highlighted in yellow and green, respectively. The peptides used in this study are represented above the aligned proteins. Regions of the cE5 protein shown by crystallographic analysis to interact with the exosite I and the catalytic site of thrombin are indicated below the alignment, as are their flanking residues. VectorBase accession numbers: A. gambiae, AGAP008004; Anopheles coluzzii, ACOM030673; Anopheles arabiensis, AARA005120; Anopheles quadriannulatus, AQUA000660; Anopheles merus, AMEM012461; Anopheles funestus, AFUN004964; Anopheles minimus, AMIN006263; Anopheles culicifacies, ACUA026308; Anopheles stephensi, ASTE006313; Anopheles dirus, ADIR011024; Anopheles sinensis, ASIS010361; A. albimanus, AALB014019. GenBankTM accession number: A. darlingi, GI:208657572. Sequences of the Anopheles melas, Anopheles christyi, A. epiroticus, Anopheles farauti, and A. atroparvus orthologues can be found in Arcà et al. (12). The sequences were aligned with Clustal Omega (43).