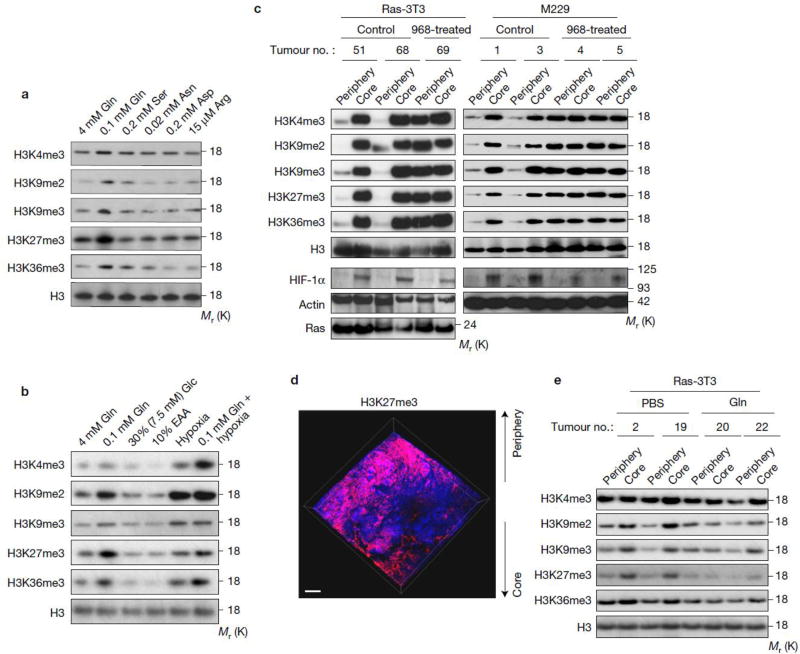
Figure 3. Glutamine deficiency drives histone hyper-methylation in tumour core regions.
(a) M229 cells were cultured in complete medium and the medium with specified concentrations (which are similar as the concentration in tumour cores) of select amino acids for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. (b) M229 cells were cultured under conditions indicated for 4 days, then cells were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. EAA, essential amino acid. Total histone H3 was used as loading control. (c) Control and 968-treated xenograft tumours were harvested. Tissues from the periphery and core regions were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 and Actin were used as loading controls. (d) 3D imaging of M229 xenograft tumour treated with 968 depicting DAPI (blue) and H3K27me3 antibody (red). Scale bar = 300 µm. (e) 4mM glutamine or PBS control injected tumours were harvested. Tissues from the periphery and core regions were lysed for histone extraction and histone lysine methylation levels were assessed by western blotting. Total histone H3 was used as loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 8.