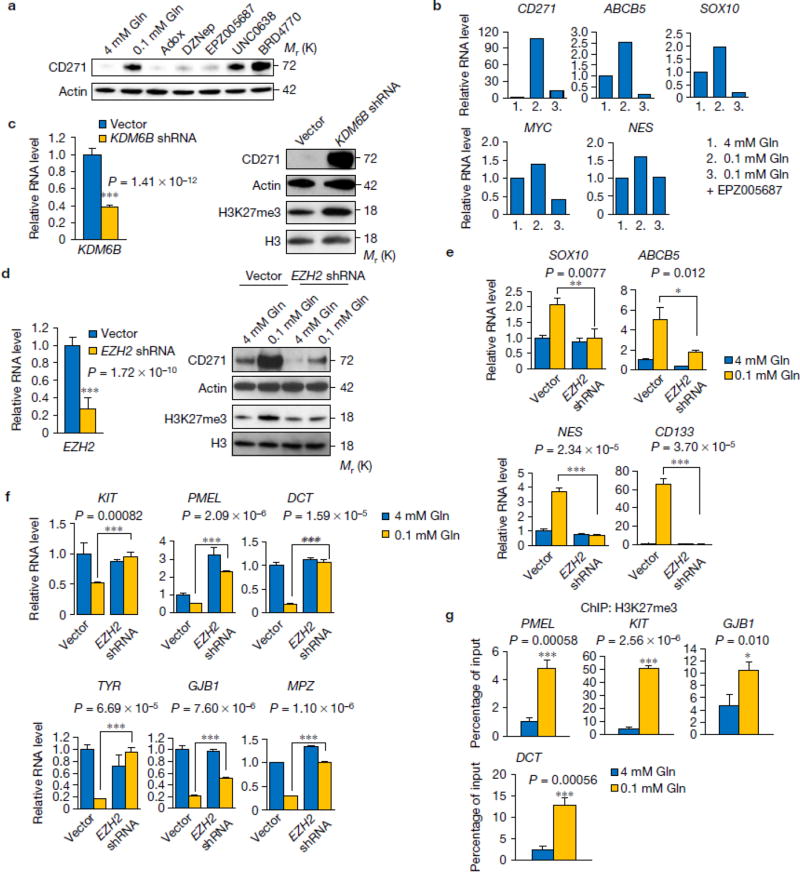
Figure 6. Low glutamine-induced de-differentiation is mediated by histone methylation on H3K27.
(a) M229 cells were cultured in complete medium or 0.1mM glutamine medium with different histone methylation inhibitors for 4 days, whole cell lysates were collected and protein levels were assessed by western blotting. Actin was used as loading control. (b) M229 cells were cultured in complete medium or 0.1mM glutamine medium with or without H3K27 specific methylation inhibitor EPZ005687 for 4 days, then RNA was extracted and de-differentiation related gene expression was assessed by qPCR. Data represent the mean of 2 biologically independent RNA extracts. Two experiments were repeated independently with similar results. Source data are shown in supplementary Table 4. (c) M229 cells were transduced with lenti viral KDM6B shRNA. Knockdown efficiency was measured by qPCR (left). Data represent mean ± S.D., n=8 biologically independent RNA extracts. ***P<.001 by unpaired two-tailed Student’s t-test. H3K27me3 and CD271 level were measured by western blotting (right). (d) M229 cells were transduced with lenti viral EZH2 shRNA. Knockdown efficiency was assessed by qPCR (left). ). Data represent mean ± S.D., n=9 biologically independent RNA extracts. ***P<.001 by unpaired two-tailed Student’s t-test. CD271 and H3K27me3 level were measured by western blotting (right). (e, f) After transduction of lenti viral EZH2 shRNA, cells were cultured in complete or 0.1mM glutamine for at least 4 days, then RNA was extracted from bulk M229 cells, de-differentiation (e) and differentiation (f) related gene expression was checked by qPCR. Data represent mean ± S.D., n=3 biologically independent RNA extracts. *P<.05, **P<.01, ***P<.001 by unpaired two-tailed Student’s t-test. (g) Cells were cultured in complete or 0.1mM glutamine medium for 4 days, then cells were harvested for ChIP assay using antibody against H3K27me3. Immunoprecipitation of the neural differentiation-related gene promoters (GJB1 and DCT in M229, PMEL and KIT in M249) were analyzed by qPCR. Data represent mean ± S.D., n=3 independent experiments. *P<.05, ***P<.001 by unpaired two-tailed Student’s t-test. Source data are shown in supplementary Table 4. Unprocessed original scans of blots are shown in Supplementary Fig. 8.