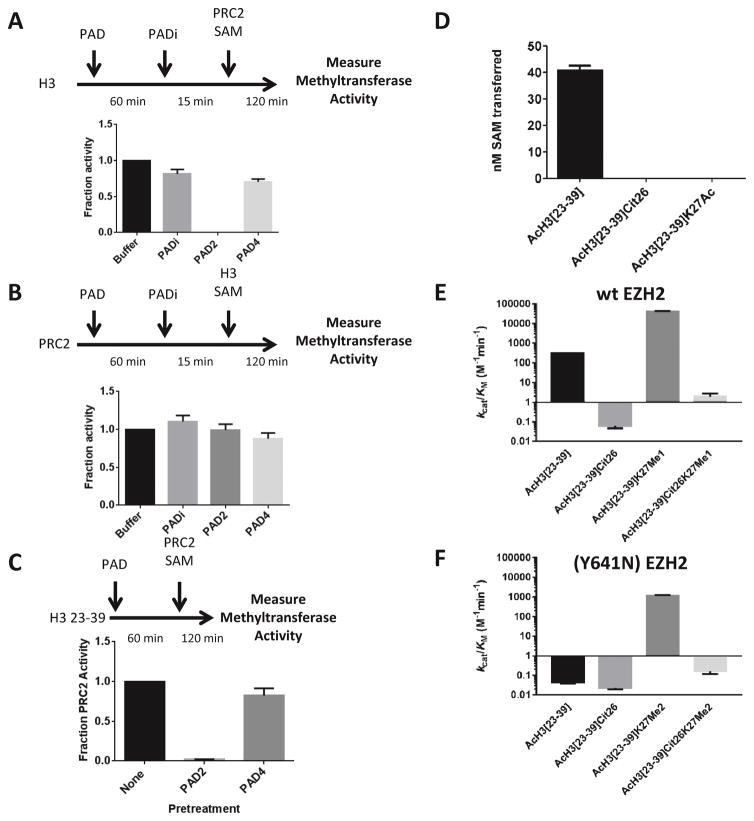
Figure 2.
PADs 2 and 4 inhibit PRC2-mediated methylation of histone H3 by modifying the histone. (A) PAD2 and PAD4 (10 nM) were preincubated with histone H3, followed by the addition of the PAD-specific inhibitor F-Amidine (1 mM). The residual methyltransferase activity was then measured using a gel-based radioactivity assay. Activity was normalized to the buffer control and is displayed as a fraction of this control. (B) PAD2 and PAD4 (10 nM) were preincubated with the PRC2 complex, followed by PAD inhibition with F-Amidine (1 mM). The measurement of residual methyltransferase activity shows that citrullination of the PRC2 complex does not impair its methyltransferase activity. (C) PAD2 and PAD4 were preincubated with a histone H3 tail analog encompassing both R26 and K27. This peptide was used because it only contains one arginine residue (R26). Thus, any effects on PRC2 activity can be attributed to the modification of R26 only. (D) A synthetic peptide containing citrulline in the place of R26 is not methylated by PRC2 complex. As a control, we show that a K27Ac containing peptide is also not methylated by the PRC2 complex. Both the Cit26 peptide as well as the K27Ac peptide showed no detectable product formation. (E,F) Methyltransferase activity of the PRC2 complex containing wt (E) or EZH2 (Y641N) (F) was measured with the indicated peptide substrate. kcat/KM is plotted to show the reduction in activity upon citrulline incorporation into the peptide substrates in the place of R26. The steady-state kinetic parameters are provided in Tables S2 and S3.