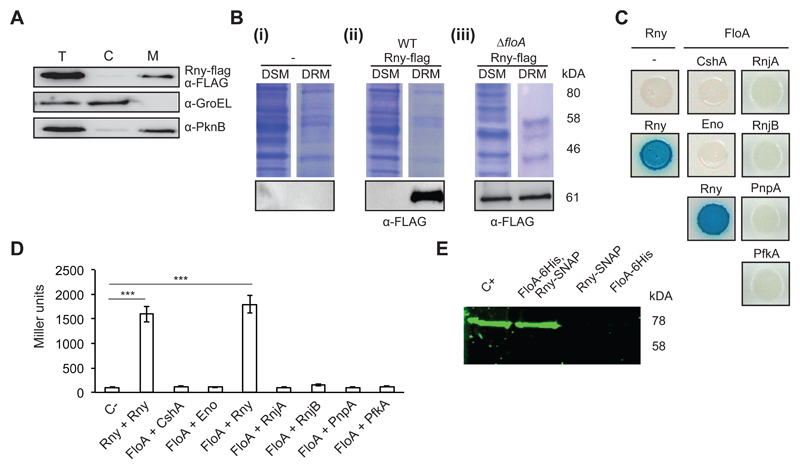
Figure 1. Rny is a membrane-bound protein associated with the DRM fraction.
(A) Immunoblot detection of Rny in the total cell (T), cytoplasmic (C) and membrane fractions (M) of Staphylococcus aureus cells using antibodies to the Flag epitope. The control for cytoplasm was an antibody to the soluble chaperone GroEL and for membrane, antibody to the membrane-bound sensor kinase PknB. (B) Immunodetection of Rny in DSM and DRM fractions of S. aureus cells. (i) Control lanes; (-) indicates an unlabeled strain. (ii) Detection of Rny in an Rny-Flag-labeled strain using anti-Flag antibodies. (iii) Detection of Rny in an Rny-Flag-labeled strain of the ΔfloA mutant. Top, Coomassie-stained gel; bottom, immunoblot. (C) Bacterial two-hybrid (BTH) analysis to study Rny and FloA interactions. (i) Interaction activates lacZ expression, leading in turn to degradation of X-Gal (blue signal). Positive control, Rny self-interaction; negative control, Escherichia coli strains bearing empty plasmids (pKNT25 + pUT18). (D) Quantification of interaction efficiency in a β-galactosidase assay; a 700 Miller Units threshold limit defines positive and negative interaction signals. Significance was measured by one-way ANOVA. p <0.005, ***. Data shown as mean ± SD of three independent biological replicates (n = 3). Each biological replicate included three technical replicates. (E) In-gel detection of Rny-SNAP using the SNAP-binding fluorescence probe TMR-Star to detect Rny in protein samples pulled down with FloA-His6. The arrow indicates band of the size predicted for Rny. Positive control (C+) is the Rny-SNAP-labeled wild-type (WT) membrane fraction. Negative controls (FloA-His6 and Rny-SNAP) were eluted from nickel-charged columns. Protein samples pulled down with FloA-His6/Rny-SNAP are shown in lanes FloA-His6 and Rny-SNAP.