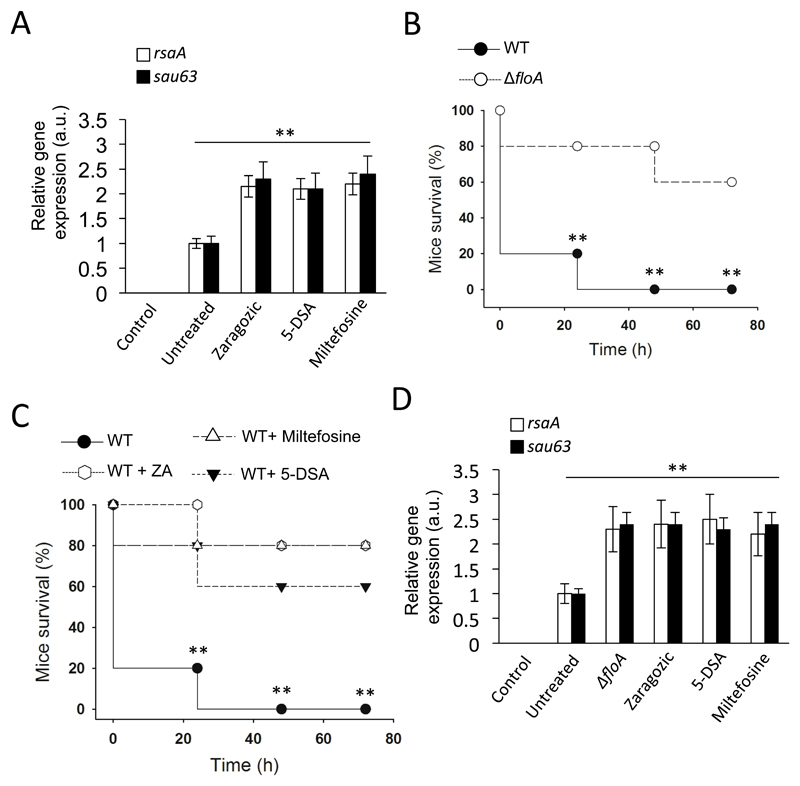
Figure 7. Inhibition of flotillin oligomerization in S. aureus attenuates virulence in a murine infection model.
(A) qRT-PCR analysis of rsaA and sau63 expression in ZA-, miltefosine- and 5-DSA-treated cells. Significance was measured by one-way ANOVA with Tukey’s comparison. For untreated vs. treated samples, ** p <0.01. Data shown as mean ± SD of three independent biological replicates (n = 3). Each biological replicate included three technical replicates. (B) Survival (%) of S. aureus-infected mice (n = 5) with WT and ΔfloA. Pneumonia caused by intranasal instillation of 3 x 108 colony-forming units in 20 μl saline solution. Infections were allowed to progress for four days. Differences in survival were analyzed by the log-rank test. Statistical significance was measured by unpaired Student's t-test, ** p <0.01. Data shown as mean ± SD of endpoint vital measurements. (C) Survival (%) of S. aureus-infected mice (n = 5), untreated or treated with ZA, milfefosine or 5-DSA (50, 20, 10 mg/kg/day respectively). Pneumonia caused by intranasal instillation of 3 x 108 colony-forming units in 20 μl saline solution. Small molecules were administered to infected mice 30 min post-infection via intraperitoneal injection. Infections were allowed to progress for four days. Differences in survival were analyzed by the log-rank test. Statistical significance was measured by unpaired Student's t-test between; Untreated vs. 5-DSA-treated; Untreated vs. ZA-treated and Untreated vs. Miltefosie-treated samples. ** p <0.01. Data shown as mean ± SD of endpoint vital measurements. (D) qRT-PCR analysis of rsaA and sau63 expression in mice. Lungs were collected 48 h post-infection and rsaA/sau63 expression levels determined. Significance was measured by one-way ANOVA with Tukey’s comparison. For untreated vs. treated samples, ** p <0.01. Data shown as mean ± SD of three independent biological replicates (n = 3). Each biological replicate included three technical replicates.