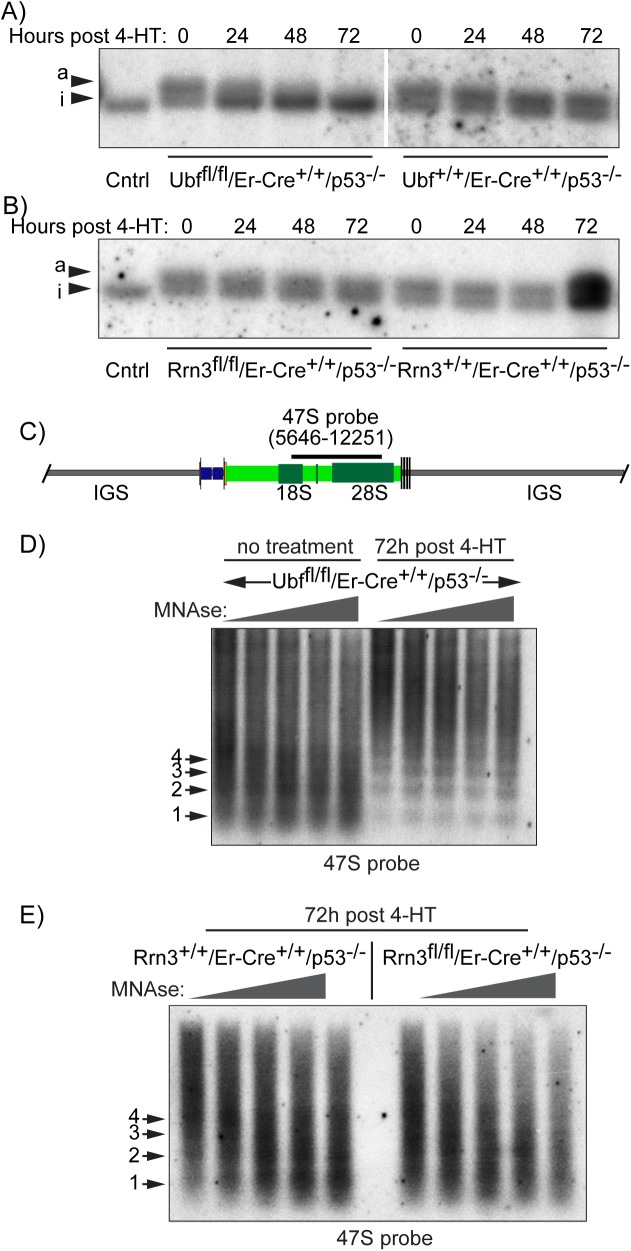
Fig 4. UBF determines the activated state of the rDNA chromatin.
A) and B) show psoralen accessibility analyses during the time course of UBF and Rrn3 gene deletion (see 4HT time course S3 Fig). The high accessibility so-called active “a” gene fraction, and the low accessibility inactive. “i” gene fraction were detected using the 47S probe. C) The mapped position of the hybridization probe relative to the start of the 47S rRNA is indicated above a diagram of the rDNA repeat. D) The profiles of increasing MNase cleavage of chromatin from UBF conditional MEFs either before or after (72h post 4-HT) inactivation of the UBF gene. E) The profiles of increasing MNase cleavage of chromatin from Rrn3 wild type and conditional (floxed) MEFs after 72h of treatment with 4-HT. The data in D) and E) was obtained using the 47S hybridization probe shown in C), and the positions of mono- (1), di- (2), etc nucleosomes are indicated. More complete datasets of the MNase analyses are given in S5 Fig.