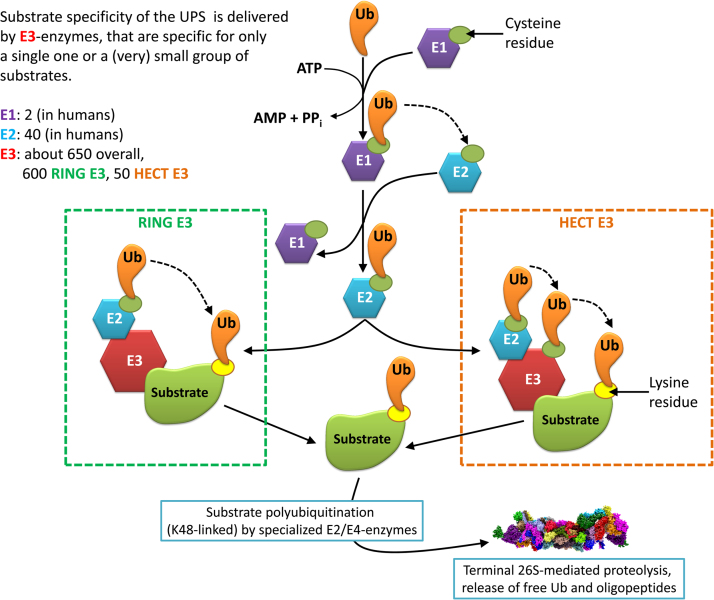
Fig. 4.
The ubiquitination pathway. This figure shows the process of substrate labeling for terminal 26S proteasomal degradation. The first step is the “activation” of an Ubiquitin (Ub) by an E1 in an ATP-consuming manner. Then Ub is transferred by E1 to an E2 enzyme. Substrate specificity is provided by the large variety of available E3 enzymes that only target a small amount of substrates. There exist two types of E3 ubiquitin ligases: RING and HECT. In case of “RING” E3: both the substrate and the Ub-loaded E2 are bound by the E3, the Ub is directly transferred from E2 to the substrate. In case of “HECT” E3, the substrate is transferred from E2 to E3 and then from E3 to the substrate. After attachment of the first Ub to the substrate, a chain of Ub-molecules is attached to the first one by specialized E2/E4-enzymes that are only able to append Ub to another Ub-molecule. The substrate is then degraded by the 26S proteasome (a chain of Ub4 provides the strongest degradation signal), the Ub-chain is released into the cytosol and monomerized again for further loading of an E1 enzyme.