Fig. 3.
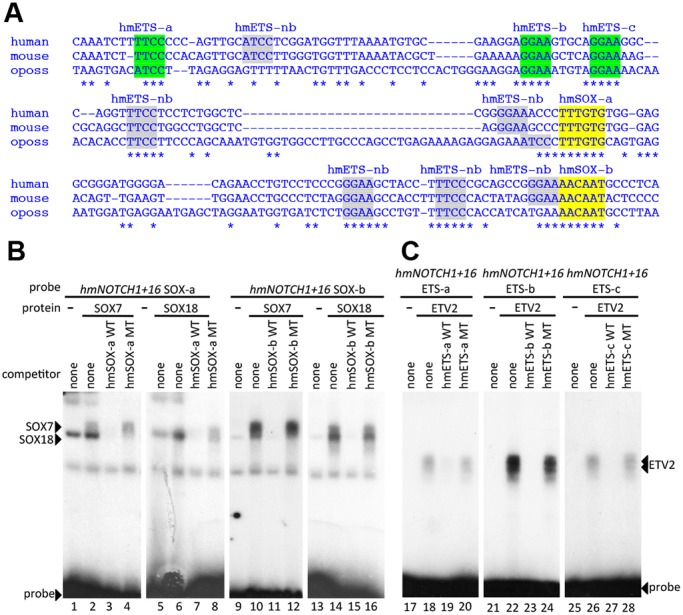
The NOTCH1+16 enhancer contains SOX and ETS binding motifs. (A) Multispecies alignment of the orthologous region of the NOTCH1+16 enhancer from human, mouse and opossum (oposs) using ClustalW. Coloured sequences are confirmed by EMSA; grey sequences are motifs identified in silico that did not bind in EMSA. (B) Radiolabelled oligonucleotide probes encompassing NOTCH1+16 hmSOX-a (lanes 1-8) and hmSOX-b (lanes 9-16) were bound to recombinant SOX7 (lanes 2-4 and 10-12) and SOX18 (lanes 6-8 and 14-16). Both proteins, which efficiently bound labelled probes (lanes 2, 6, 10 and 14), were competed by excess unlabelled self-probe (WT, lanes 3, 7, 11 and 15) but not by mutant self-probe (MT, lanes 4, 8, 12 and 16). (C) Radiolabelled oligonucleotide probes encompassing NOTCH1+16 hmETS-a (lanes 17-20), hmETS-b (lanes 21-24) and hmETS-c (lanes 25-28) were bound to recombinant ETV2 protein. ETV2, which efficiently bound to labelled probes (lanes 18, 22 and 26), was competed by excess unlabelled self-probe (WT, lanes 19, 23 and 27) but not by mutant self-probe (MT, lanes 20, 24 and 28).
