Abstract
The transition from proliferation to specification is fundamental to the development of appropriately patterned tissues. In the developing Drosophila eye, Eyes absent (Eya) and Sine oculis (So) orchestrate the progression of progenitor cells from asynchronous cell division to G1 arrest and neuronal specification at the morphogenetic furrow. Here, we uncover a novel role for Eya and So in promoting cell cycle exit in the second mitotic wave (SMW), a synchronized, terminal cell division that occurs several hours after passage of the furrow. We show that Combgap (Cg), a zinc-finger transcription factor, antagonizes Eya-So function in the SMW. Based on the ability of Cg to attenuate Eya-So transcriptional output in vivo and in cultured cells and on meta analysis of their chromatin occupancy profiles, we speculate that Cg limits Eya-So activation of select target genes posterior to the furrow to ensure properly timed mitotic exit. Our work supports a model in which context-specific modulation of transcriptional activity enables Eya and So to promote both entry into and exit from the cell cycle in a distinct spatiotemporal sequence.
KEY WORDS: Eye development, Cell cycle, Transcription factor, Retinal determination gene network, Mitotic exit
Summary: Combgap acts as a novel negative regulator of Eya-So in transcriptional outputs that coordinate cell cycle exit with the transition to differentiation during Drosophila retinal development.
INTRODUCTION
Development of a functional organ requires coordination of proliferation with specification, differentiation, and morphogenesis. The capacity to divide typically inversely correlates with the progress of a cell toward terminal differentiation, such that mitotic cells are unspecified whereas differentiating cells have permanently exited the cell cycle (Buttitta and Edgar, 2007; Zhu and Skoultchi, 2001). Although many of the signaling pathways and transcription factors that direct these transitions have been identified, how inputs that promote and inhibit the cell cycle are balanced to schedule mitosis according to context is poorly understood (Brown et al., 2003; Buttitta and Edgar, 2007; Devès and Bourrat, 2012; Zhu and Skoultchi, 2001).
The unique spatiotemporal pattern of proliferation and differentiation in the Drosophila eye imaginal disc makes it an ideal model with which to study the underlying regulation. Prior to the third larval instar, asynchronous proliferation generates the pool of progenitor cells from which the different retinal cell types will be specified (Kumar, 2011). Differentiation begins when a burst of Decapentaplegic (Dpp) and Hedgehog (Hh) signaling at the posterior margin arrests cells in G1 phase of the cell cycle and initiates the morphogenetic furrow (MF) (Chanut and Heberlein, 1997; Curtiss and Mlodzik, 2000; Dominguez and Hafen, 1997; Firth and Baker, 2005; Greenwood and Struhl, 1999; Horsfield et al., 1998; Ready et al., 1976; Vrailas and Moses, 2006; Wolff and Ready, 1991). As the MF propagates anteriorly across the eye field, arrested progenitor cells are either recruited into photoreceptor preclusters and specified as R8, R2, R5, R3 and R4 neurons or undergo one final synchronized round of mitosis known as the second mitotic wave (SMW) (Wolff and Ready, 1991). Progenitors that divide in the SMW adopt either an R1, R6 or R7 photoreceptor fate or the ommatidial accessory cone, pigment, and bristle fates (Wolff and Ready, 1991).
The retinal determination gene network (RDGN), which is composed of the transcription factors Eyeless (Ey), Eyes absent (Eya), Sine oculis (So) and Dachshund (Dac), governs the transition between proliferation and specification that occurs around the MF (Kumar, 2009). Anterior to the MF, coexpression of Ey with the transcription factors Homothorax (Hth), Yorkie and Teashirt promotes proliferation of the unspecified retinal precursors (Bessa et al., 2002; Lopes and Casares, 2010; Peng et al., 2009). Induction of Dpp and Hh signaling then represses Hth, enabling the onset of Dac, Eya and So expression (Bessa et al., 2002; Chen et al., 1999; Curtiss and Mlodzik, 2000; Halder et al., 1998; Lopes and Casares, 2010; Niimi et al., 1999; Pappu, 2003; Pappu et al., 2005; Salzer and Kumar, 2009). Dac interferes with the pro-proliferative transcriptional complexes to terminate mitogenic activity and permit differentiation, while Eya and So, operating as a bipartite transcription factor, take control of the cell cycle by activating a burst of string (stg) transcription that forces cells into M phase and prepares them for coordinated G1 arrest (Brás-Pereira et al., 2015; Jemc and Rebay, 2007; Pignoni et al., 1997). Shortly thereafter, Eya-So first cooperates with Ey to activate atonal expression and initiate neuronal specification and then directly represses ey transcription to stabilize the suppression of the earlier proliferation program in the differentiating cells (Atkins et al., 2013; Zhang et al., 2006; Zhou et al., 2014).
After recruitment of the photoreceptor preclusters, a new combination of signaling and transcription factor inputs reinitiates the cell cycle, directing the remaining progenitors to undergo a single, synchronized and final division. Dpp, Hh, Notch and EGFR signaling all promote SMW proliferation, with antagonistic regulation of stg expression by the transcriptional activator and EGFR effector Pointed (Pnt) and the transcriptional repressor Tramtrack (Ttk) controlling the timing of M-phase entry (Baker and Yu, 2001; Baonza and Freeman, 2005; Baonza et al., 2002; Duman-Scheel et al., 2002; Firth and Baker, 2005, 2007; Sukhanova and Du, 2008; Vrailas and Moses, 2006; Yang and Baker, 2003, 2006). Whether these are the only SMW regulators and how their activities are tuned to ensure that most unspecified precursors divide once with the correct timing is unknown and, in particular, the role of Eya-So has not yet been explored.
Here, we present evidence that antagonism between Eya-So and the transcription factor Combgap (Cg) helps to direct SMW proliferation in the Drosophila retina. Our data suggest that Eya-So stabilizes cell cycle exit to limit the number of mitoses undertaken by precursor cells in the SMW. By contrast, Cg promotes SMW proliferation such that its loss leads to a failure to generate sufficient precursors to assemble the complete eye. Mutual genetic antagonism between Eya-So and Cg suggests that their interaction contributes to the balance between positive and negative inputs that restricts precursor cells to a single mitosis after the MF passes. Mechanistically, we propose that Cg opposes Eya-So transcriptional activity, as Cg limits the ability of Eya-So to activate target genes both in vivo and in S2 cell-based transcription assays. Together, our results identify the first negative regulator of Eya-So transcriptional activity, uncover a novel context in which Eya-So controls the cell cycle, and bolster the model that balanced positive and negative transcriptional inputs calibrate the rate of SMW proliferation.
RESULTS
Eya-So limits SMW proliferation
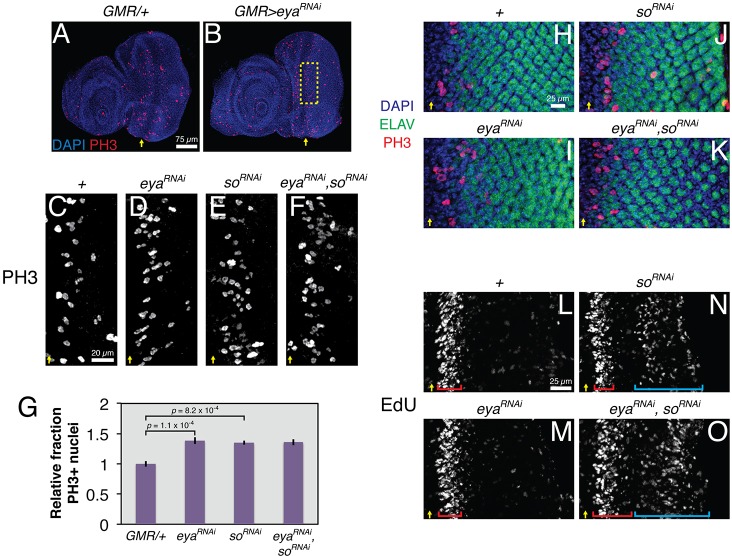
Although Eya-So is known to contribute to the control of proliferation and cell cycle arrest at the onset of the MF (Jemc and Rebay, 2007; Karandikar et al., 2014; Lopes and Casares, 2015), its role in regulating the subsequent coordinated precursor cell mitosis known as the SMW has not been investigated. Because null clones of eya or so block MF progression (Pignoni et al., 1997), precluding analysis of the SMW, we instead used GMR-GAL4 to express UAS-RNAi transgenes targeting their transcripts in all cells posterior to the MF. eyaRNAi strongly knocked down its target, as judged by a >85% reduction in Eya protein levels when driven with GMR-GAL4 and a ∼65% penetrant ʻeyeless' phenotype (with only small patches of retinal tissue in the remaining 35%) when driven with ey-GAL4 (Fig. S1 and see Fig. 2G). Using anti-phosphorylated histone H3 (PH3) to mark mitotic cells, we found that knocking down eya or so, or both together, increased the rate of SMW mitosis by almost 50% (Fig. 1A-G). The lack of PH3 signal in ELAV-positive photoreceptors, together with the normal complement of ELAV-positive cells in the preclusters near the SMW, suggested that the extra PH3-positive nuclei resulted from progenitors undergoing extra mitoses, rather than from specified cells re-entering the cell cycle (Fig. 1H-K). The number of PH3-positive nuclei posterior to the SMW region did not increase (Fig. 1H-K). Consistent with eya operating as an SMW antagonist, reducing the dose of positive versus negative regulators of M phase suppressed and enhanced the GMR>eyaRNAi phenotype, respectively (Fig. S2). We conclude that Eya-So participates in the transcriptional events that regulate mitosis at the SMW.
Fig. 2.
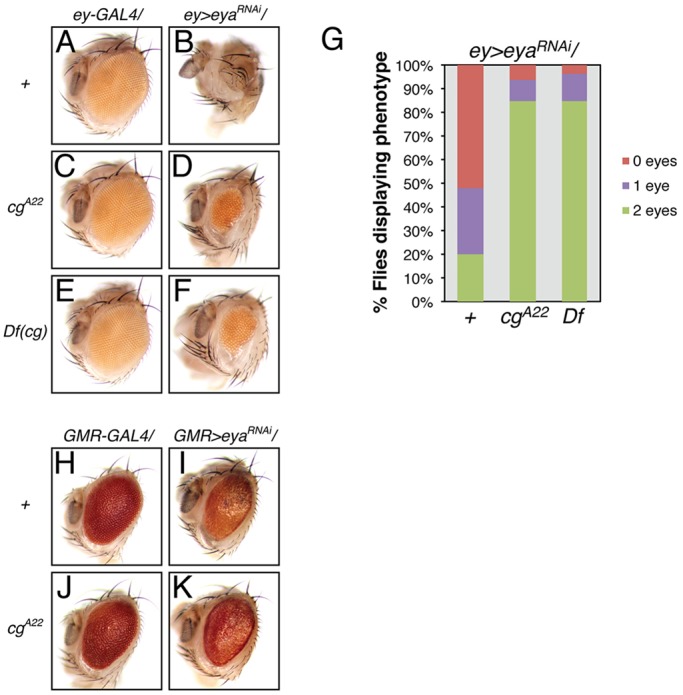
cg antagonizes eya functions during retinal development. (A,B) Adult ‘eyeless' phenotype associated with ey-GAL4-driven eya knockdown; eye loss is never observed in the driver-alone control. (C-F) cg heterozygosity, with either a null allele or a deficiency, suppresses the ey>eyaRNAi eyeless phenotype. (G) Quantification of penetrance of eye loss (n≥91 animals). Flies were scored for the presence of pigmented retinal tissue in both eye fields. (H,I) GMR-GAL4-driven eya knockdown reduces eye size and pigmentation; the driver alone produces a mild rough eye phenotype. (J,K) cg heterozygosity restores eye size and pigmentation in eya knockdown animals, but does not modify the driver-alone phenotype.
Fig. 1.
Eya-So limits mitosis in the SMW. All images show representative late third instar eye-antennal imaginal discs. Yellow arrows mark the ventral edge of the MF. UAS transgenes were expressed with GMR-GAL4. (A,B) eya knockdown increases the number of PH3+ nuclei in the SMW relative to the driver-alone control. (C-F) Magnified views of the SMW (boxed region in B) show increased numbers of PH3+ nuclei in eya and so knockdown discs relative to driver-alone controls. Double knockdown does not further increase SMW mitoses. (G) Quantification of mitotic rates for genotypes in C-F (n≥5), calculated as the number of PH3+ nuclei per μm of the MF and normalized to controls. (H-K) Inter-ommatidial spacing, marked by ELAV− DAPI-stained nuclei, is reduced and irregular in eya or so knockdown discs. In the double knockdown, a reduced number of ELAV+ photoreceptors in the posterior produces the appearance of increased spacing. PH3+ ELAV+ nuclei were never detected. (L-O) EdU incorporation shows that so knockdown induces ectopic S phases (cyan brackets) posterior to the SMW (red brackets). eya knockdown enhances this phenotype.
Next, we determined which phase of the cell cycle Eya-So regulates. Knocking down eya or so alone did not increase EdU incorporation at the SMW (Fig. 1L-N). However, depleting both expanded the domain of SMW cells in S phase (compare red brackets, Fig. 1L-O). Furthermore, knocking down so initiated ectopic S phases in cells posterior to the SMW, and expressing eyaRNAi exacerbated this phenotype (cyan brackets, Fig. 1N,O). The loss of ELAV-positive cells posterior to the SMW in eyaRNAi, soRNAi discs (Fig. 1K), together with the increased EdU incorporation relative to that in the so knockdown alone (Fig. 1N,O), suggests that failure to stabilize the post-mitotic state of newly specified photoreceptors might contribute to the aberrant S-phase re-entry observed in double-knockdown retinas. Thus, Eya-So limits both S-phase and M-phase entry or progression and is required for cell cycle exit posterior to the MF.
Cg, a new eya-interacting transcription factor, limits Eya-So transcriptional output
That Eya is a transcriptional coactivator for So is well established (Ohto et al., 1999; Silver et al., 2003; Xu et al., 1997), and recent work has shown that Eya-So directly represses ey expression in differentiating cells posterior to the MF (Atkins et al., 2013). Extrapolating to the context of the SMW, Eya-So could limit proliferation by either activating the expression of negative regulators or repressing the expression of positive regulators, with different outputs presumably determined by its co-factors. However, of the few Eya-So binding partners studied to date, none is implicated in SMW regulation (Ahmed et al., 2012; Chen et al., 1997; Goldstein et al., 2005; Morillo et al., 2012; Pignoni et al., 1997). With the goals of expanding the repertoire of Eya-interacting factors and gaining additional insight into SMW regulation, we carried out a yeast two-hybrid screen using the conserved C-terminal Eya domain (ED) from Drosophila Eya as bait. Positive hits were tested for their ability to dominantly modify eya loss-of-function phenotypes anterior or posterior to the MF.
Among the most significant two-hybrid hits was a collection of clones whose overlap defined a span of eight zinc-fingers within the coding sequence of the transcription factor Cg. In follow-up genetic tests, heterozygosity for a cg null allele dominantly suppressed eya loss-of-function phenotypes. Specifically, in an ey>eyaRNAi hypomorphic background in which only 20% of adults developed bilateral eye tissue, reduction in cg dose increased that fraction to 80% (Fig. 2A-G), whereas in the GMR>eyaRNAi background it ameliorated the reduced pigmentation and increased adult eye size (Fig. 2H-K). These suppressive interactions classified cg as a genetic antagonist of eya both anterior and posterior to the MF.
cg was identified by Calvin Bridges and named to describe its roles in limiting the number of male sex combs and in patterning the wing veins (Lindsley and Zimm, 1992). Despite its ubiquitous expression in the eye-antennal imaginal disc (Fig. S3), its contributions to retinal development have not been described. Mechanistically, Cg works as a sequence-specific DNA-binding protein that contributes to recruitment of Polycomb repressive complexes (Ray et al., 2016), can organize the three-dimensional conformation of target loci (Hitrik et al., 2016), and is thought to activate and repress targets in the leg, wing and brain (Campbell and Tomlinson, 2000; Song et al., 2000; Svendsen et al., 2000). Based on this prior knowledge, Cg presented an intriguing candidate for a transcription factor that might influence the output of Eya-So.
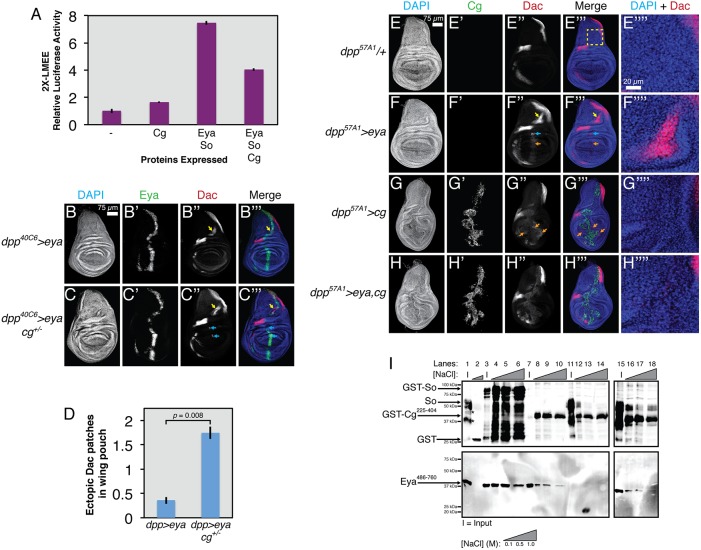
To test this possibility, we asked whether Cg could alter Eya-So activity in an S2 cell-based transcription assay using an Eya-So-responsive transcriptional reporter carrying two copies of the lozenge minimal enhancer element (LMEE) (Yan et al., 2003) upstream of a minimal promoter and the coding sequence of firefly luciferase. Consistent with prior work (Mutsuddi et al., 2005; Silver et al., 2003), transient transfection of eya and so increased luciferase activity almost eightfold compared with the control (Fig. 3A). Co-transfection of cg with eya and so reduced reporter expression by half, a decrease made even more significant considering the slight activation observed when cg was transfected alone. Eya and So expression and subcellular localization were not affected by Cg overexpression, with all three proteins colocalizing in S2 cell nuclei (Fig. S4). The ability of Cg to reduce Eya-So activity was specific, as Cg did not reduce output from the transcriptional activator Pnt in similar assays (Fig. S5).
Fig. 3.
Cg antagonizes transcriptional output from Eya-So. All stained tissues are late third instar wing imaginal discs (n>10 discs examined for all genotypes). (A) Cg limits the ability of Eya-So to activate LMEE-luciferase in S2 cell transcription assays (seven independent replicates). (B) dpp-GAL4-driven eya misexpression always induces ectopic Dac expression in the hinge (yellow arrows, compare with E″), but only occasionally in the pouch (not shown). (C) cg heterozygosity enhances the ability of eya to activate ectopic Dac in the pouch (cyan arrows). (D) Quantification of the number of ectopic Dac patches in the pouch for genotypes in B,C (n≥8). (E-H) A stronger dpp-GAL4 driver. (E″″-H″″) Magnified views of the dorsal patch of ectopic Dac, as boxed in E‴. (E) Driver alone control showing the wild-type pattern of Dac expression. (F) eya misexpression induces ectopic Dac in the hinge (yellow arrow), pouch (blue arrow) and faintly throughout the dpp stripe in the pouch (orange arrow). (G) cg overexpression produces smaller wing discs with aberrant folds in the pouch and loss of the normal dorsal-ventral asymmetric shape and induces weak Dac expression in the pouch, both cell-autonomously and non-autonomously (orange arrows; note lack of overlap with Cg-expressing cells in the dpp stripe). (H) Wing discs co-overexpressing cg and eya no longer ectopically induce Dac in the hinge and pouch and display milder morphological disruptions than with cg overexpression alone. (I) In vitro GST pulldown assays show that Cg can directly bind Eya and can participate in a ternary complex with Eya-So (six independent replicates). Lanes 1-2, GST alone does not pull down So or FLAG-Eya486-760. Full-length So runs as a triplet and the asterisk marks co-purifying degradation products. Lanes 3-6, GST-So pulls down FLAG-Eya486-760. Lanes 7-10, GST-Cg225-404 pulls down FLAG-Eya486-760. Lanes 11-14, GST-Cg225-404 does not effectively pull down So. Lanes 15-18, FLAG-Eya486-760 increases the ability of GST-Cg225-404 to pull down So.
To examine whether Cg could antagonize Eya-So transcriptional output in vivo, we turned to a wing misexpression assay that has been used extensively to reveal regulatory relationships and to validate transcriptional targets of the RDGN (Chen et al., 1997; Halder et al., 1998; Jemc and Rebay, 2007; Morillo et al., 2012). In this experiment, eya misexpression along the anterior-posterior compartment boundary of the larval wing imaginal disc under the control of dpp-GAL4 ectopically activates expression of the Eya-So target gene dac in a so-dependent manner. Cg is expressed uniformly throughout the wing disc (Campbell and Tomlinson, 2000; Svendsen et al., 2000), so we predicted that if Cg limits Eya-So transcriptional activity, then decreasing or increasing the cg dose should respectively enhance or reduce the ability of Eya to activate ectopic Dac expression. As previously reported (Morillo et al., 2012), eya misexpression under dpp40C6-GAL4 always induced a large dorsal patch of ectopic Dac (yellow arrow, Fig. 3B) and, at lower frequency, small clusters of Dac-positive cells in the wing pouch near the hinge (cyan arrows, compare Fig. 3B,C with E). Driving eya with a slightly stronger GAL4 transgene (dpp57A1-GAL4) also activated weak Dac expression throughout the dpp domain (orange arrow, Fig. 3F). cg heterozygosity in the dpp40C6>eya background increased the number of clusters of wing pouch Dac-positive cells fourfold, suggesting that endogenous cg limits the ability of Eya-So to activate dac transcription (Fig. 3B-D). Consistent with this idea, co-overexpressing cg and eya eliminated induction of ectopic Dac in the dorsal wing or pouch, but did not alter the low-level Dac induced in the dpp domain (Fig. 3H). Eya protein levels and subcellular localization were unchanged by altering cg dose (Fig. S6), indicating that the antagonism of Eya-mediated Dac induction by Cg most likely reflects reduced Eya-So transcriptional activity.
In the course of the cg overexpression experiments, we noticed that dpp>cg alone disrupted wing disc morphology such that the discs were smaller, with aberrant folding patterns and partial loss of the physical asymmetry between anterior and posterior halves (Fig. 3G). Cg overexpression also reduced the ventral and medial patterns of endogenous Dac expression, presumably reflecting the altered morphology and fate of tissue in those regions. Weak, sometimes non-autonomous Dac misexpression was also observed in the anterior and posterior pouch compartments in these discs (orange arrows, Fig. 3G). Co-overexpression of eya with cg reduced the severity of cg-induced disruptions to overall disc morphology (Fig. 3H, Fig. S6), hinting that not only does Cg antagonize the transcriptional function of Eya, but also that Eya might reciprocally inhibit Cg.
Because we initially identified Cg as an Eya binding partner, we used in vitro pulldown assays with bacterially expressed proteins to test two different molecular models for how Cg might reduce Eya-So transcriptional activity. First, Eya bound to Cg might no longer interact with So. Alternatively, Cg might form a ternary complex with Eya-So that is less transcriptionally active than Eya-So alone. Because full-length Eya and Cg were poorly expressed and were largely insoluble and degraded (data not shown), we used smaller fragments that corresponded to the putative interaction domains identified by the two-hybrid screen. Confirming the two-hybrid result, GST-Cg225-404 pulled down Eya486-760, whereas GST alone did not (Fig. 3I, compare lanes 8-10 with lane 2). When GST-Cg225-404 was mixed with full-length So, only an extremely weak interaction was detected (lanes 12-14). Addition of Eya486-760 enabled GST-Cg225-404 to pull down So, revealing formation of a ternary complex in which Eya bridges the interaction (lanes 16-18). We attempted to corroborate this result by co-immunoprecipitating the full-length proteins from transiently transfected S2 cells, but failed to detect the Eya-Cg interaction (data not shown), suggesting that Eya-Cg complexes might be more labile in cells than in vitro.
Cg promotes SMW proliferation and Eya-So opposes this activity
Returning to the developing retina, we examined whether cg genetically antagonizes eya function in the context of the SMW. Indeed, cg heterozygosity dominantly suppressed the increased rate of SMW mitoses produced by GMR>eyaRNAi knockdown almost back to the wild-type level (Fig. 4A-C,G). Conversely, cg overexpression, which on its own produced a mild increase in PH3-positive SMW cells, enhanced the eya knockdown phenotype (Fig. 4D,E,H). Over the course of several experiments, only two animals in the latter experiment (Fig. 4D,H) and four overexpressing cg alone (Fig. 4E,H) survived to the late third instar, making statistical analyses difficult. However, the SMW proliferation rate of both eyaRNAi,cg larvae exceeded the expected result if GMR>eyaRNAi and GMR>cg additively interacted, supporting a model of genetic antagonism (2.16-fold and 2.61-fold increases; additive expectation 1.92-fold). These results both pinpoint the SMW as a developmental context in which eya and cg interact and suggest that cg positively regulates proliferation posterior to the MF.
Fig. 4.
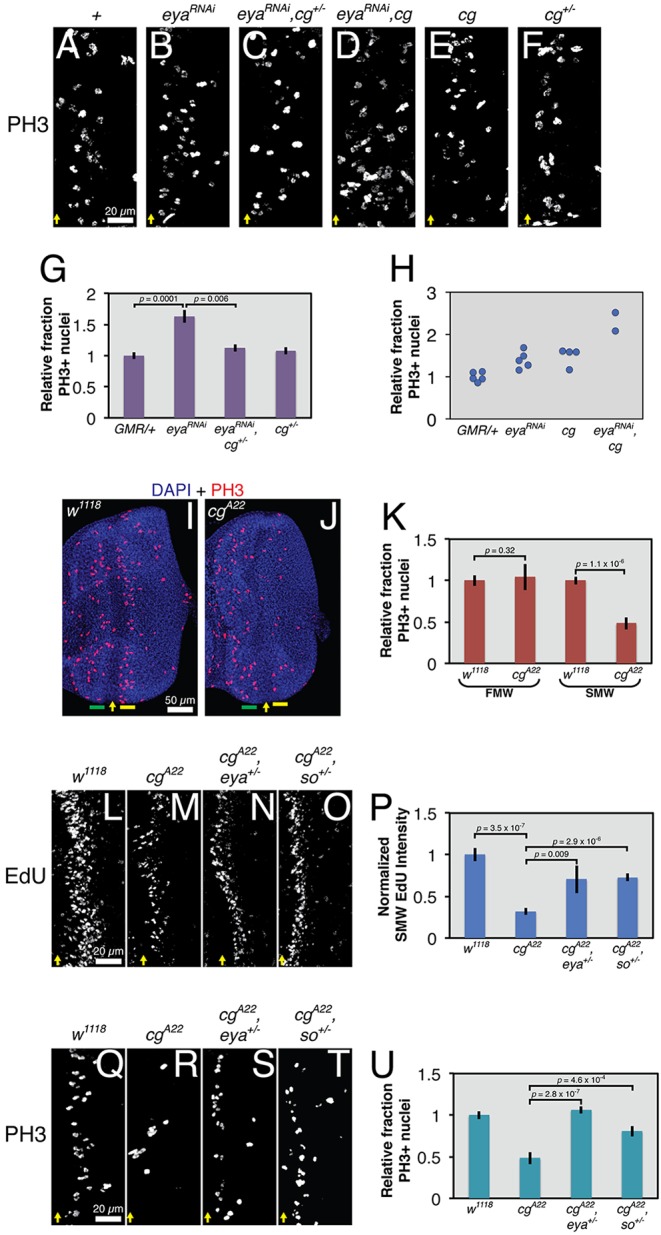
Antagonistic inputs from cg and eya regulate SMW proliferation. All images show representative late third instar eye-antennal imaginal discs. Yellow arrows mark the ventral edge of the MF. UAS transgenes were expressed with GMR-GAL4. (A-D) cg heterozygosity suppresses and cg overexpression enhances the elevated SMW mitotic rate of eya knockdown animals. (E,F) cg overexpression modestly increases SMW proliferation, whereas heterozygosity has no effect. (G) Quantification of data in A,B,C,F (n≥5), calculated as the number of PH3+ nuclei per μm of the MF and normalized to controls. (H) Quantification of data in A,B,D,E, calculated as in G. (I,J) Relative to control, cg loss reduces the number of PH3+ nuclei in the SMW (yellow bar) but not in the first mitotic wave (FMW, green bar). (K) Quantification of data in I,J (n≥9), calculated as in G. (L-O) cg null discs incorporate less EdU at the SMW than wild-type controls, and eya or so heterozygosity dominantly suppresses this phenotype. (P) Quantification of background-subtracted fluorescent intensity of SMW EdU signal, normalized to the control intensity, for genotypes in L-O (n≥5). (Q-T) eya or so heterozygosity dominantly suppresses the reduction in PH3+ nuclei in the SMW in cg mutants. (U) Quantification of data in Q-T (n≥11), calculated as in G. Actual PH3+ counts for A-F and Q-T are provided in Table S1.
To confirm the requirement of cg for the SMW, we examined the consequences of complete cg loss. As predicted, the SMW mitotic rate of cg null discs was half that of wild-type controls (Fig. 4I-K). Proliferation anterior to the MF (the first mitotic wave, or FMW) was unchanged, indicating that the requirement of cg for mitosis is specific to the SMW. Furthermore, cg mutants incorporated ∼75% less EdU at the SMW than wild-type animals, suggesting that cg is required for efficient S-phase entry or progression (Fig. 4L,M,P). Heterozygosity for eya or so suppressed both the reduced EdU incorporation and lower proportion of PH3-positive nuclei in the SMW of cg null discs (Fig. 4L-U). These results suggest that the mutually antagonistic relationship between cg and eya-so helps set the correct SMW proliferation rate.
cg is required for normal eye-antennal disc morphology and cell survival
In addition to SMW phenotypes, we noted other defects in cg mutant eye-antennal discs. First, although neuronal specification appeared largely normal in cg null retinas as judged by the pattern of the marker ELAV (Fig. 5A,B), MF progression was delayed in the dorsal quarter such that the MF curved toward the posterior in this region (Fig. 5B,D,F, white arrows). This phenotype was variably penetrant in its severity, but the MF curvature consistently differed from that of the wild type (compare Fig. 5A,C,E with 5B,D,F). Second, an ectopic flap of tissue protruding from between the developing eye and antenna extended basally and posteriorly (Fig. 5B,D,F, red arrows). This flap was variable in size and sometimes reached underneath the MF, but we never observed defects in MF progression or differentiation that spatially correlated with apposition of the flap and retinal cells. The regional identity of the ectopic flap might be presumptive head cuticle based its location and expression of markers such as Cut (Fig. 5C-F) (Blochlinger et al., 1993; Weasner and Kumar, 2013). Third, cg mutant antennal discs frequently displayed a narrowed and misshapen antennal ring (Fig. 5C-F). Finally, cg loss induced abnormal apoptosis early in the third instar, with the greatest amount of cell death concentrated between the antenna and eye near the dorsal-ventral boundary, just anterior to the MF near the dorsal and ventral margins, and in the anterior of the antenna (Fig. 5G,H). This wave of apoptosis was largely complete by the late third instar. Thus, in addition to controlling SMW proliferation, cg appears to have roles in directing epithelial morphogenesis, stabilizing regional identities, and promoting cell survival.
Fig. 5.
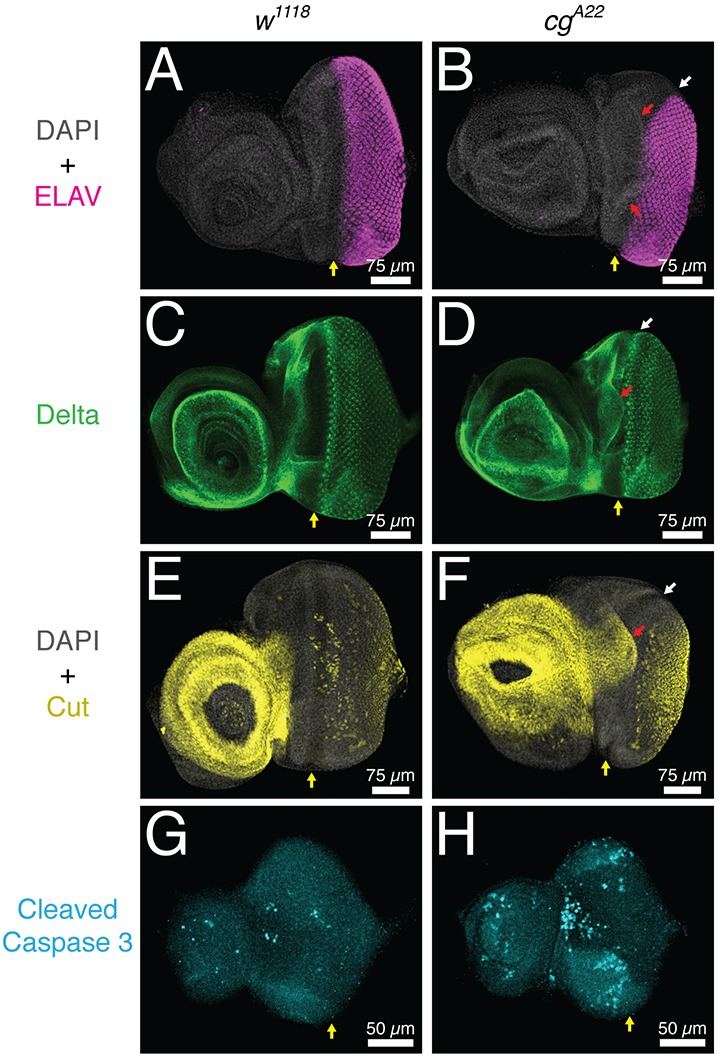
cg is required for larval eye-antennal imaginal disc morphogenesis. All images show representative third instar eye-antennal imaginal discs. Yellow arrows mark the ventral edge of the MF. (A-F) Dorsal MF progression appears delayed (white arrows), presumptive head tissue is expanded (red arrows), and the overall morphology and folding of the antennal disc is irregular in late third instar cg mutants. (A,B) Photoreceptor specification, marked by ELAV expression, appears normal in cg mutants. (C,D) Delta is expressed in the expanded flap of tissue (red arrow) but otherwise appears normal in cg mutants. (E,F) Ectopic Cut expression is observed in the expanded flap of tissue (red arrow). Specification of Cut-positive cone cells appears normal in cg mutants. (G,H) Although overall cg disc morphology appears normal at the early third instar, increased apoptosis is detected in the antennal disc, at the antennal-eye boundary in the region where the extra flap of tissue will form, and in the lateral margins of the eye disc just anterior to the MF.
Cell types generated by the SMW are progressively missing in cg mutants
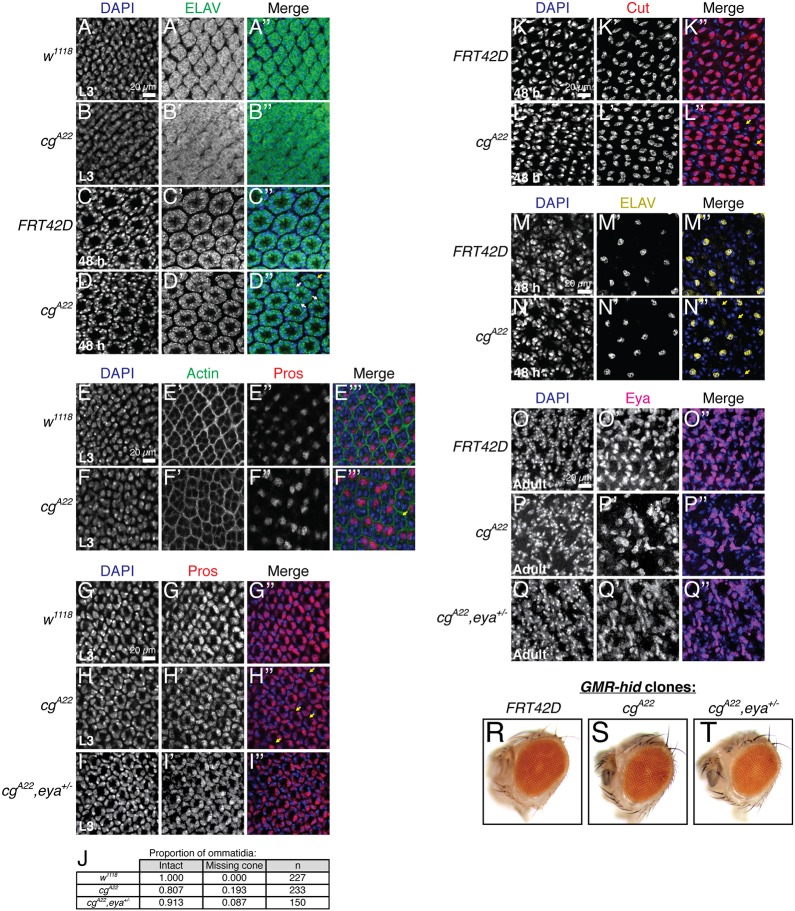
The SMW generates a precursor pool sufficient for recruitment of photoreceptors 1, 6 and 7, cone cells, pigment cells and bristle cells. Thus, insufficient mitoses should cause loss of these retinal cell types, with the latest specified cells most significantly impacted as the precursors are depleted. To assess the complement of cell types present in cg null retinal tissue, we stained with markers of various cell fates at several developmental time points. For analysis at pupal stages, we bypassed the larval lethality of cgA22 homozygotes with the cell lethal/GMR-hid system (Stowers and Schwarz, 1999) to generate retinas composed entirely of cg null tissue. First, recruitment of the ELAV-positive photoreceptors appeared nearly normal in cg mutants, with ommatidia only occasionally lacking a photoreceptor (Fig. 6A-D, yellow arrow). However, the ommatidia were packed more closely together and mildly disorganized in these third instar discs (Fig. 6A,B). Ommatidial organization appeared almost normal in the pupal GMR-hid cg clones, although occasionally the inter-ommatidial spacing was reduced, consistent with reduced SMW proliferation producing a smaller progenitor pool (Fig. 6A-D, white arrows). Second, and consistent with the ELAV results, Prospero (Pros)-positive R7 photoreceptors, which are recruited after the SMW, were rarely missing (Fig. 6E,F, yellow arrow), although their spacing and organization were abnormal. Third, ∼20% of third instar cg mutant ommatidia had only three cone cells, as compared with the four present in wild type (Fig. 6G,H,J, yellow arrows), a frequency of loss consistent with the range reported for other mutants that reduce the SMW (Du et al., 1996). Focusing on the apical plane where the cone cell nuclei reside, ommatidial organization appeared essentially normal in the cg third instar disc (Fig. 6H). This observation suggests that irregular packing of the newly specified photoreceptor clusters (Fig. 6B′) might be a temporary aberration caused by proliferation defects and disorganization within the basal progenitor pool; restoration of a close to normal ommatidial lattice in the cg pupal disc (Fig. 6D′) is consistent with this idea. Fourth, cg mutant eyes contained fewer ELAV-positive bristle cells (Fig. 6M,N, yellow arrows) and Eya-positive pigment cells (Fig. 6O,P), supporting a progressive model of loss. Finally, cg whole-eye clones were smaller and rougher than their control counterparts (Fig. 6R,S), consistent with descriptions of mutants that ablate the SMW (de Nooij and Hariharan, 1995; Du et al., 1996; Xin et al., 2002).
Fig. 6.
Cell types specified after the SMW are progressively missing in cg null retinal tissue. All images show confocal projections of five or fewer slices at the apical-basal position of each cell type. When cell loss is claimed, complete confocal projections were examined to ensure that the cell in question was not mislocalized rather than absent. Third instar discs (L3) are from null cg mutants, while cg clones were used for pupal and adult time points. (A,B) Reduced inter-ommatidial space produces a disorganized ommatidial lattice in cg null third instar eye discs. (C,D) Patterning appears more regular in 48 h pupal whole-eye cg clones, although occasional reduction in inter-ommatidial distance (white arrows) and missing ELAV+ photoreceptors (yellow arrow) are observed (D″). (E,F) Pros+ R7 photoreceptors are irregularly spaced (F″) and occasionally missing (yellow arrow, F‴) in cg null L3 retinas. (G-I) Ommatidia with only three Pros+ cone cells are frequently observed in cg null L3 eye discs (yellow arrows, H″) and eya heterozygosity suppresses this phenotype. (J) Quantification of the number of ommatidia containing the normal complement of cone cells (intact) or missing one cone cell for the genotypes in G-I. (K,L) Ommatidia with fewer Cut+ cone cells are also observed in 48 h pupal cg null clones (yellow arrows, L″). (M,N) 48 h pupal cg null ommatidia sometimes lack ELAV+ bristle cells (yellow arrows, N″). (O-Q) Significant loss and disorganization of the Eya+ inter-ommatidial pigment cell lattice in adult cg clones as compared with control clones. Halving eya dosage suppresses pigment cell loss. (R-T) eya heterozygosity suppresses the mild rough eye phenotype of cg mutant retinal tissue.
Based on these analyses, we propose that cg mutant tissue, because of reduced SMW proliferation, might not generate adequate retinal precursors from which to assemble complete ommatidia. However, an alternate possibility is that the loss of cell fates is not connected to the reduced SMW proliferation, but rather reflects independent roles for cg in specifying or maintaining late-born cell fates. To distinguish these models, we focused on the cone cells. First, if cg were required to maintain cone cell fate, then we expected progressive loss of expression of cone cell markers. Although cg mutant ommatidia sometimes lacked a cone cell in the 48 h pupal disc (Fig. 6K,L, yellow arrows), the frequency of loss was not greater than in the third instar disc, arguing against a requirement for cg in cell fate maintenance. Second, we reasoned that because eya promotes cone cell specification (Karandikar et al., 2014), if the loss of cone cells in the cg mutant reflected an independent role in the signaling events that specify this fate, then heterozygosity for eya should enhance that phenotype. By contrast, if the reduced number of cone cells in the cg mutant stems from an insufficient progenitor pool, then because eya heterozygosity restored SMW proliferation in the cg mutant (Fig. 4L-U) it should also suppress the cone cell specification defect. Consistent with the latter prediction, eya heterozygosity suppressed cone cell loss in cg null discs (Fig. 6H-J). Reducing eya dosage also ameliorated pigment cell loss (Fig. 6P,Q) and adult eye roughness (Fig. 6S,T) in cg mutant tissue. Together, our results argue that insufficient SMW proliferation in the cg mutant compromises the ability to assemble complete ommatidia and confirm the requirement for eya-cg genetic antagonism in promoting normal retinal development.
Genes bound by both So and Cg are enriched for SMW regulators
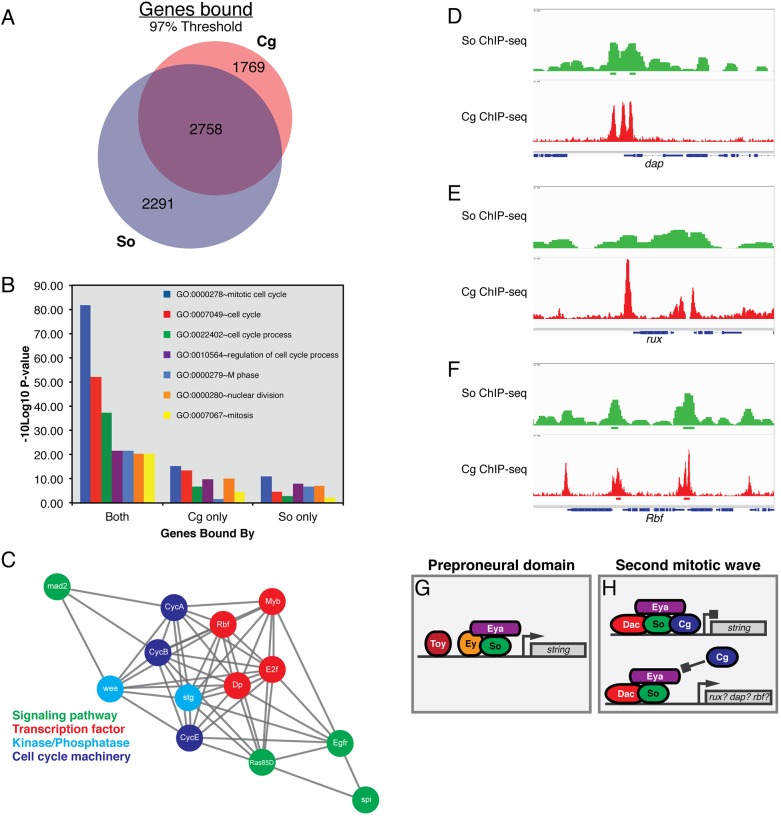
Our finding that Cg can participate in the Eya-So complex in vitro and inhibit the ability of Eya-So to promote transcription motivated us to compare the published So and Cg genome-wide ChIP-seq profiles derived from third instar imaginal discs (Jusiak et al., 2014a,b; Ray et al., 2016). This analysis revealed that 61% of genes bound by Cg within 3 kb of their transcription start sites also contained a So peak in the same interval; conversely, Cg occupied 55% of genes bound by So (Fig. 7A). Co-occupied genes were enriched for gene ontology terms reflecting cell cycle regulation, as compared with loci bound by either protein alone (Fig. 7B). Of particular interest, a substantial population of genes experimentally shown to direct the SMW cell cycle were bound by both So and Cg (Fig. 7C). Consistent with a model in which Eya-So supports cell cycle exit by activating the transcription of cell cycle inhibitors, and Cg interferes with this regulation, we noted overlapping So and Cg occupancy at dacapo (dap), roughex (rux) and Retinoblastoma-family protein (Rbf) (Fig. 7D-F), although at subthreshold levels at the former two loci. Rbf is the most promising candidate, as eya overexpression has been reported to increase its transcription threefold (Jemc and Rebay, 2007) (Fig. S7). Collectively, these analyses are consistent with the hypothesis that Eya-So and Cg antagonistically control SMW proliferation by co-occupying and regulating the expression of genes that control this cell cycle.
Fig. 7.
Overlapping chromatin occupancy of So and Cg at genes that regulate the SMW. (A) The proportion of genes bound by So, Cg, or both at a 97% threshold. (B) Gene ontology (GO) terms associated with the cell cycle are enriched for the set of genes bound by both So and Cg. (C) Genes that control the SMW cell cycle are bound by So and Cg. Gray lines signify protein-protein, genetic, or computationally predicted interactions between genes as annotated by STRING. (D-F) So and Cg occupancy at dap, rux and Rbf. Peak calls are shown below the occupancy profiles. Peaks at dap and rux fell just under our stringent threshold. (G,H) RDGN transcription factors might assemble heterogeneous complexes at target loci to differentially regulate the cell cycle anterior and posterior to the morphogenetic furrow. ‘Preproneural domain' refers to cells immediately anterior to the furrow, while ‘second mitotic wave' denotes precursor cells undergoing their terminal division posterior to the furrow.
DISCUSSION
Here we present evidence that Eya-So promotes exit of retinal precursors from the cell cycle after their final mitosis to permit their recruitment into ommatidia. Combined with published data, this finding suggests that the RDGN initiates both timely entry into and exit out of the cell cycle as retinal cells prepare to differentiate (Bessa et al., 2002; Brás-Pereira et al., 2015, 2016; Jemc and Rebay, 2007; Lopes and Casares, 2015). We also identify Cg as a novel positive regulator of the decision to re-enter S phase after G1 arrest in the MF, and consequently expand the repertoire of transcription factors whose balanced positive and negative inputs link cell cycle progression with retinal specification. The ability of Cg to curb Eya-So transcriptional output, coupled with mutual genetic antagonism between these genes at the SMW, suggests that the dynamic assembly of uniquely composed Eya-So complexes with specific transcriptional activities coordinates proliferation with other cellular decisions in the developing eye.
Our finding that Eya-So limits proliferation in the SMW suggests that regulation of the cell cycle by this transcriptional complex is more elaborate in two respects than is currently assumed. First, Eya-So does not promote proliferation in all contexts. Most prior studies conducted in flies and mammals have concluded that Eya and So family proteins stimulate proliferation during normal development and tumorigenesis. For example, Eya and Six proteins promote progression into S phase by activating transcription of the Cyclin A1 or Cyclin D1 genes (Coletta et al., 2004; Hua et al., 2014; Li et al., 2003, 2013), while Drosophila Eya-So stimulates M-phase entry by activating stg transcription (Jemc and Rebay, 2007). However, a recent study found that eya null cells in the MF inappropriately maintain Cyclin B expression, reflecting failure to arrest in G1 (Karandikar et al., 2014), while we show that SMW cells with reduced Eya-So expression undergo extra mitoses, indicating that they cannot exit the cell cycle with the correct timing in preparation for terminal differentiation. Second, Eya-So does not control the cell cycle exclusively at the G1-S and G2-M transitions. Our experiments reveal a novel third node of regulation by Eya-So: stabilization of cell cycle exit upon completion of M phase. Knocking down eya and so posterior to the MF induced ectopic DNA synthesis but not inappropriate M-phase entry, suggesting both that Eya-So prevents these precursors from proceeding into S phase and that the signaling and transcriptional environment once cells leave the SMW does not support progression further into the cell cycle. Many posterior cells lost ELAV expression in this experiment, consistent with either failed recruitment into ommatidia or an inability to stabilize neuronal fate upon re-entry into the cell cycle. We favor the latter interpretation, as neither eya nor so is required for larval photoreceptor differentiation (Jin et al., 2016).
These insights augment the model that the RDGN choreographs the sequence of cell cycle and developmental events that generates an eye. Early in retinal development, Ey cooperates with Hth and Teashirt to promote asynchronous proliferation (Bessa et al., 2002). Just before the MF initiates, another RDGN member, Dac, inhibits Hth-containing transcriptional complexes to terminate the early proliferation program, while Eya-So, Ey and Twin of Ey (Toy) activate stg transcription to push cells that have passed G1 through M phase (Brás-Pereira et al., 2015; Jemc and Rebay, 2007; Lopes and Casares, 2015). A burst of Dpp and Hh signaling then synchronously arrests cells in the MF at G1 (Escudero and Freeman, 2007; Firth and Baker, 2005; Horsfield et al., 1998; Vrailas and Moses, 2006). Dac collaborates with Dpp and Hh to initiate cell cycle exit at the MF (Brás-Pereira et al., 2015, 2016), and Eya also contributes (Karandikar et al., 2014), but it is unclear what molecular switch terminates Eya-So activation of stg transcription and balances the positive and negative inputs of retinal determination proteins to achieve this event. Transcriptional co-factors, such as Cg, are likely to tune Eya-So transcriptional activity, as we discuss below. Once the MF passes, Ey and Eya-So cooperate to promote atonal transcription and initiate neurogenesis (Zhang et al., 2006; Zhou et al., 2014). Our work attributes a final point of RDGN regulation of the cell cycle to Eya-So, which we show limits the remaining unspecified precursor cells in the SMW to a single division and maintains cell cycle arrest thereafter.
Illuminating the mechanisms that tailor the magnitude and direction of the effect of the RDGN on the cell cycle to specific developmental time points is an outstanding challenge. One emerging hypothesis is that Eya-So switches between activating and repressing its transcriptional targets, but the co-factors that drive these shifts and their relevance to proliferation are unknown. Specifically, So promotes ey transcription in retinal precursors, while Eya-So inhibits its expression in differentiating cells (Atkins et al., 2013). Similarly, Eya-So activates stg expression anterior to the MF, but not when cells arrest in G1 (Jemc and Rebay, 2007). The inhibition by Cg of Eya-So transcriptional output in vivo and its participation in the complex in vitro make Cg a tantalizing candidate co-repressor that might terminate activation of transcription by Eya-So or initiate active repression to orchestrate the cell cycle. For example, Eya-So could contribute to cell cycle exit posterior to the MF by initiating transcription of genes that interfere with S-phase re-entry, such as Rbf, rux or dap (Buttitta et al., 2007; De Nooij et al., 1996; Lane et al., 1996; Ruggiero et al., 2012), but Cg might impede activation at these loci to permit cell cycle re-entry as cells enter the SMW. The enrichment of Cg and So co-occupancy at genes that govern proliferation (Fig. 7B-F), along with our finding that cg antagonizes the ability of eya-so to promote cell cycle exit, supports this hypothesis. The recent identification of Cg as a DNA-binding transcription factor that can recruit chromatin-remodeling Polycomb group (PcG) proteins (Ray et al., 2016) offers a potential repressive mechanism, and the specific links between epigenetic regulation and the RDGN are compelling subjects for future study.
While we favor a model in which Cg limits Eya-So function posterior to the MF and contributes to the transition from pro-mitotic activity to anti-proliferative function as the MF passes, all three proteins are broadly expressed in the larval retina (Fig. S3), indicating that some additional input must dictate this switch (Bonini et al., 1993; Campbell and Tomlinson, 2000; Cheyette et al., 1994; Svendsen et al., 2000). Intriguing candidates may be found among other retinal determination transcription factors. Ey and Toy coactivate stg transcription with Eya-So anterior to the MF (Jemc and Rebay, 2007; Lopes and Casares, 2015), but are not expressed at the SMW (Czerny et al., 1999; Halder et al., 1998). By contrast, Dac interferes with proliferation anterior to the MF (Brás-Pereira et al., 2015) and is required for cell cycle arrest after the SMW (Brás-Pereira et al., 2016). Therefore, perhaps Eya-So assembles activating transcriptional complexes with Ey and Toy at stg that override repressive Cg input in the preproneural domain, but interacts with Cg and Dac to inhibit stg transcription and maintain cell cycle exit posterior to the MF (Fig. 7G,H). Consistent with the hypothesis that Eya-So represses stg after the SMW, so knockdown ectopically activates a stg enhancer that is normally expressed only in the anterior (Lopes and Casares, 2015). Given the biochemical evidence for both Eya-So and Eya-Dac complexes (Chen et al., 1997; Jin and Mardon, 2016; Mutsuddi et al., 2005; Pignoni et al., 1997; Zhang et al., 2006), differential stg regulation anterior and posterior to the MF could reflect assembly of heterogeneous RDGN transcriptional complexes. However, posterior ey misexpression did not alter the rate of mitosis in the SMW (data not shown), indicating that Ey alone is not sufficient to reverse the direction of the effect of Eya-So on proliferation. Moreover, ey was not derepressed in cg clones posterior to the MF, and cg overexpression anterior to the MF did not repress ey (Fig. S8), arguing that Cg does not control proliferation by influencing Eya-So regulation at ey. Elucidating how the combinatorial action of unique RDGN protein complexes dictates enhancer specificity at stg and other loci to schedule cell cycle events is a rich topic for future study.
MATERIALS AND METHODS
Drosophila strains
UAS-eyaRNAi and UAS-soRNAi were obtained from the Vienna Drosophila Resource Center. The following were from the Bloomington Drosophila Stock Center: GMR-GAL4Zipursky, pntΔ88, ttk1e11, ey-GAL4, Df(2R)BSC401/CyO, dpp40C6-GAL4, eyaClift, so3, ey-FLP;FRT42D,ubi-GFP/CyO, y1,w*;FRT42D,y+,GMR-hid,l(2)CL-R1/Cyo;ey-GAL4,UAS-FLP and FRT42D. Additional strains: cgA22 (a gift of Gerard Campbell, University of Pittsburgh, PA, USA), UAS-stgRNAi (Kyoto Stock Center), dpp57A1-GAL4 (Staehling-Hampton et al., 1994), UAS-eyaI (Hsiao et al., 2001), UAS-eyaIIIa (Hsiao et al., 2001) and UAS-cg (this work). For further details of Drosophila strains and genetics, see the supplementary Materials and Methods.
Immunohistochemistry and microscopy
Imaging was performed with a Zeiss LSM 510 or Zeiss LSM 880 confocal microscope, using 0.5 to 1.0 μm steps and projecting maximally through the desired tissue unless otherwise noted. See supplementary Materials and Methods for the antibodies used. P-values were calculated using two-tailed Student's t-tests of two-sample equal variance between the indicated genotypes, and data are plotted as mean±s.e.m. To image adult eyes, 3- to 5-day-old adult flies were decapitated and photographed with a Canon EOS Rebel camera fitted to a Leica dissecting microscope. Individual slices were merged using iSolution-Lite software (IMT-Digital).
In vitro pulldown assays
The Cg225-404 fragment was PCR amplified from cg with 5′-ATAGGATCCCTGCTGCTTCACTCCACGGAGAGACC-3′ and 5′-AAACTCGAGGTTGGGATTGACGCCATTGTGCG-3′ and ligated into pGEX-4T-1. GST, So and Eya recombinant proteins were prepared and the pulldown assays performed as previously described (Morillo et al., 2012). See the supplementary Materials and Methods for details.
Transcription assays
2.25×106 Drosophila S2 cells plated in 12-well plates were transfected with 750 ng total plasmid DNA as described in the supplementary Materials and Methods. cg and eya were expressed constitutively under the Actin5C promoter, while so expression was controlled with the CuSO4-inducible Metallothionein promoter (plasmid pRmHA-3; Silver et al., 2003).
Bioinformatic analysis of So and Cg ChIP-seq datasets
We obtained the genomic coordinates of the top 3% of peaks by height using the Integrated Genome Browser (Freese et al., 2016); this threshold is more stringent than either published set of peak calls (Jusiak et al., 2014a,b; Ray et al., 2016). A list of genomic coordinates ±3 kb from every transcription start site in the genome (assembly dm3) was generated using bedtools slop and merged with peak lists using bedtools intersect to produce lists of transcription start sites associated with peaks of So only, Cg only, or both So and Cg. Fig. 7A was generated using BioVenn (Hulsen et al., 2008), gene ontology terms were analyzed with the DAVID Functional Annotation tool (Huang et al., 2008, 2009), the protein-protein interaction network of genes co-bound by So and Cg was generated using STRING (Szklarczyk et al., 2015), and peaks were visualized with the Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013).
Acknowledgements
We thank Gerard Campbell for FRT42D,cgA22 flies; Judith Kassis for sharing her lab's called Cg peaks; Jemma Webber, Nicelio Sanchez-Luege and Matt Hope for help with bioinformatics; Aaron Mitchell-Dick for initial exploration of yeast two-hybrid hits; members of the Justin Kumar, Wei Du and David Kovar labs for advice on the EdU protocol and pulldown experiments; Rick Fehon for the Delta and cleaved caspase 3 antibodies and the equipment to photograph adult eyes; current and former members of the I.R. and Fehon labs, Chip Ferguson, David Kovar, Doug Bishop and Jim Holaska for helpful discussions; and Matt Hope and Nicelio Sanchez-Luege for critiquing this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: I.R., T.L.D.; Methodology: I.R., T.L.D.; Validation: I.R., T.L.D.; Formal analysis: T.L.D.; Investigation: T.L.D.; Writing - original draft: I.R., T.L.D.; Writing - review & editing: I.R., T.L.D.; Visualization: I.R., T.L.D.; Supervision: I.R.; Project administration: I.R., T.L.D.; Funding acquisition: I.R.
Funding
This work was supported by National Institutes of Health (NIH) grant R01 EY12549 to I.R. and by the Genomics Core Facility through a University of Chicago Cancer Center Support Grant P30 CA014599. T.L.D. was supported by NIH T32 HD055164. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147231.supplemental
References
- Ahmed M., Wong E. Y. M., Sun J., Xu J., Wang F. and Xu P.-X. (2012). Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377-390. 10.1016/j.devcel.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins M., Jiang Y., Sansores-Garcia L., Jusiak B., Halder G. and Mardon G. (2013). Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet. 9, e1003731 10.1371/journal.pgen.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. and Yu S.-Y. (2001). The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104, 699-708. 10.1016/S0092-8674(01)00266-5 [DOI] [PubMed] [Google Scholar]
- Baonza A. and Freeman M. (2005). Control of cell proliferation in the Drosophila eye by notch signaling. Dev. Cell 8, 529-539. 10.1016/j.devcel.2005.01.019 [DOI] [PubMed] [Google Scholar]
- Baonza A., Murawsky C. M., Travers A. A. and Freeman M. (2002). Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4, 976-980. 10.1038/ncb887 [DOI] [PubMed] [Google Scholar]
- Bessa J., Gebelein B., Pichaud F., Casares F. and Mann R. S. (2002). Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 16, 2415-2427. 10.1101/gad.1009002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer R., Jan L. Y. and Jan Y. N. (1990). Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4, 1322-1331. 10.1101/gad.4.8.1322 [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Jan L. Y. and Jan Y. N. (1993). Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development 450, 441-450. [DOI] [PubMed] [Google Scholar]
- Bonini N. M., Leiserson W. M. and Benzer S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379-395. 10.1016/0092-8674(93)90115-7 [DOI] [PubMed] [Google Scholar]
- Brás-Pereira C., Casares F. and Janody F. (2015). The retinal determination gene dachshund restricts cell proliferation by limiting the activity of the Homothorax-Yorkie complex. Development 142, 1470-1479. 10.1242/dev.113340 [DOI] [PubMed] [Google Scholar]
- Brás-Pereira C., Potier D., Jacobs J., Aerts S., Casares F. and Janody F. (2016). dachshund potentiates hedgehog signaling during Drosophila retinogenesis. PLoS Genet. 12, e1006204 10.1371/journal.pgen.1006204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G., Hughes P. J. and Michell R. H. (2003). Cell differentiation and proliferation–simultaneous but independent? Exp. Cell Res. 291, 282-288. 10.1016/S0014-4827(03)00393-8 [DOI] [PubMed] [Google Scholar]
- Buttitta L. A. and Edgar B. A. (2007). Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697-704. 10.1016/j.ceb.2007.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta L. A., Katzaroff A. J., Perez C. L., de la Cruz A. and Edgar B. A. (2007). A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 12, 631-643. 10.1016/j.devcel.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Campbell G. L. and Tomlinson A. (2000). Transcriptional regulation of the Hedgehog effector CI by the zinc-finger gene combgap. Development 127, 4095-4103. [DOI] [PubMed] [Google Scholar]
- Campbell G., Göring H., Lin T., Spana E., Andersson S. and Doe C. Q. (1994). RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development 120, 2957-2966. [DOI] [PubMed] [Google Scholar]
- Chanut F. and Heberlein U. (1997). Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development 124, 559-567. [DOI] [PubMed] [Google Scholar]
- Chen R., Amoui M., Zhang Z. and Mardon G. (1997). Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91, 893-903. 10.1016/S0092-8674(00)80481-X [DOI] [PubMed] [Google Scholar]
- Chen R., Halder G., Zhang Z. and Mardon G. (1999). Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development 126, 935-943. [DOI] [PubMed] [Google Scholar]
- Cheyette B. N. R., Green P. J., Martin K., Garren H., Hartenstein V. and Zipursky S. L. (1994). The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron 12, 977-996. 10.1016/0896-6273(94)90308-5 [DOI] [PubMed] [Google Scholar]
- Coletta R. D., Christensen K., Reichenberger K. J., Lamb J., Micomonaco D., Huang L., Wolf D. M., Müller-Tidow C., Golub T. R., Kawakami K. et al. (2004). The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl. Acad. Sci. USA 101, 6478-6483. 10.1073/pnas.0401139101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J. and Mlodzik M. (2000). Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 1336, 1325-1336. [DOI] [PubMed] [Google Scholar]
- Czerny T., Halder G., Kloter U., Souabni A., Gehring W. J. and Busslinger M. (1999). Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell 3, 297-307. 10.1016/S1097-2765(00)80457-8 [DOI] [PubMed] [Google Scholar]
- de Nooij J. C. and Hariharan I. K. (1995). Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science 270, 983-985. 10.1126/science.270.5238.983 [DOI] [PubMed] [Google Scholar]
- de Nooij J. C., Letendre M. A. and Hariharan I. K. (1996). A cyclin-dependent kinase inhibitor, dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87, 1237-1247. 10.1016/S0092-8674(00)81819-X [DOI] [PubMed] [Google Scholar]
- Devès M. and Bourrat F. (2012). Transcriptional mechanisms of developmental cell cycle arrest: problems and models. Semin. Cell Dev. Biol. 23, 290-297. 10.1016/j.semcdb.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Dominguez M. and Hafen E. (1997). Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 11, 3254-3264. 10.1101/gad.11.23.3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W. E. I., Vidal M., Xie J.-E. and Dyson N. (1996). RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10, 1206-1218. 10.1101/gad.10.10.1206 [DOI] [PubMed] [Google Scholar]
- Duman-Scheel M., Weng L., Xin S. and Du W. (2002). Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 417, 299-304. 10.1038/417299a [DOI] [PubMed] [Google Scholar]
- Escudero L. M. and Freeman M. (2007). Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev. Biol. 13, 1-13. 10.1186/1471-213X-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., Muskavitch M. A. T. and Artavanis-Tsakonas S. (1990). Molecular interactions between the protein products of the neurogenic loci notch and Delta, two EGF-homologous genes in Drosaphila. Cell 61, 523-534. 10.1016/0092-8674(90)90534-L [DOI] [PubMed] [Google Scholar]
- Firth L. C. and Baker N. E. (2005). Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8, 541-551. 10.1016/j.devcel.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Firth L. C. and Baker N. E. (2007). Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev. Biol. 307, 521-538. 10.1016/j.ydbio.2007.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese N. H., Norris D. C. and Loraine A. E. (2016). Integrated genome browser: visual analytics platform for genomics. Bioinformatics 32, 2089-2095. 10.1093/bioinformatics/btw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R. E., Cook O., Dinur T., Pisanté A., Karandikar U. C., Bidwai A. and Pisante A. (2005). An eh1-like motif in odd-skipped mediates recruitment of groucho and repression in vivo. Mol. Cell. Biol. 25, 10711-10720. 10.1128/MCB.25.24.10711-10720.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood S. and Struhl G. (1999). Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126, 5795-5808. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Flister S., Walldorf U., Kloter U. and Gehring W. J. (1998). Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125, 2181-2191. [DOI] [PubMed] [Google Scholar]
- Hitrik A., Popliker M., Gancz D., Mukamel Z., Lifshitz A., Schwartzman O., Tanay A. and Gilboa L. (2016). Combgap promotes ovarian niche development and chromatin association of EcR-binding regions in BR-C. PLoS Genet. 12, e1006330 10.1371/journal.pgen.1006330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsfield J., Penton A., Secombe J., Hoffman F. M. and Richardson H. (1998). decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development 125, 5069-5078. [DOI] [PubMed] [Google Scholar]
- Hsiao F. C., Williams A., Davies E. L. and Rebay I. (2001). Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev. Cell 1, 51-61. 10.1016/S1534-5807(01)00011-9 [DOI] [PubMed] [Google Scholar]
- Hua L., Fan L., Aichun W., Yongjin Z., Qingqing C. and Xiaojian W. (2014). Inhibition of Six1 promotes apoptosis, suppresses proliferation, and migration of osteosarcoma cells. Tumor Biol. 35, 1925-1931. 10.1007/s13277-013-1258-1 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J. and Alkema W. (2008). BioVenn -- a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 10.1186/1471-2164-9-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemc J. and Rebay I. (2007). Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 310, 416-429. 10.1016/j.ydbio.2007.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M. and Mardon G. (2016). Distinct biochemical activities of eyes absent during drosophila eye development. Sci. Rep. 6, 23228 10.1038/srep23228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Eblimit A., Pulikkathara M., Corr S., Chen R. and Mardon G. (2016). Conditional knockout of retinal determination genes in differentiating cells in Drosophila. FEBS J. 283, 2754-2766. 10.1111/febs.13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusiak B., Karandikar U. C., Kwak S.-J., Wang F., Wang H., Chen R. and Mardon G. (2014a). Regulation of Drosophila eye development by the transcription factor Sine oculis. PLoS ONE 9, e89695 10.1371/journal.pone.0089695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusiak B., Wang F., Karandikar U. C., Kwak S.-J., Wang H., Chen R. and Mardon G. (2014b). Genome-wide DNA binding pattern of the homeodomain transcription factor Sine oculis (So) in the developing eye of Drosophila melanogaster. Genomics Data 2, 153-155. 10.1016/j.gdata.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandikar U. C., Jin M., Jusiak B., Kwak S. J., Chen R. and Mardon G. (2014). Drosophila eyes absent is required for normal cone and pigment cell development. PLoS ONE 9, e102143 10.1371/journal.pone.0102143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J. P. (2009). The molecular circuitry governing retinal determination. Biochim. Biophys. Acta 1789, 306-314. 10.1016/j.bbagrm.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J. P. (2011). My what big eyes you have: how the drosophila retina grows. Dev. Neurobiol. 71, 1133-1152. 10.1002/dneu.20921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. E., Sauer K., Wallace K., Jan Y. N., Lehner C. F. and Vaessin H. (1996). Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87, 1225-1235. 10.1016/S0092-8674(00)81818-8 [DOI] [PubMed] [Google Scholar]
- Li X., Oghi K. A., Zhang J., Krones A., Bush K. T., Glass C. K., Nigam S. K., Aggarwal A. K., Maas R., Rose D. W. et al. (2003). Eya protein phosphatase activity regulates Six1 – Dach – Eya transcriptional effects in mammalian organogenesis. Nature 426, 247-254. 10.1038/nature02083 [DOI] [PubMed] [Google Scholar]
- Li Z., Tian T., Lv F., Chang Y., Wang X., Zhang L., Li X., Li L., Ma W., Wu J. et al. (2013). Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS ONE 8, e59203 10.1371/journal.pone.0059203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D. L. and Zimm G. G. (1992). The Genome of Drosophila melanogaster. Cambridge: Academic Press. [Google Scholar]
- Lopes C. S. and Casares F. (2010). hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev. Biol. 339, 78-88. 10.1016/j.ydbio.2009.12.020 [DOI] [PubMed] [Google Scholar]
- Lopes C. S. and Casares F. (2015). Eye selector logic for a coordinated cell cycle exit. PLoS Genet. 11, e1004981 10.1371/journal.pgen.1004981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon G., Solomon N. M. and Rubin G. M. (1994). dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120, 3473-3486. [DOI] [PubMed] [Google Scholar]
- Morillo S. A., Braid L. R., Verheyen E. M. and Rebay I. (2012). Nemo phosphorylates Eyes absent and enhances output from the Eya-Sine oculis transcriptional complex during Drosophila retinal determination. Dev. Biol. 365, 267-276. 10.1016/j.ydbio.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuddi M., Chaffee B., Cassidy J., Silver S. J., Tootle T. L. and Rebay I. (2005). Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics 170, 687-695. 10.1534/genetics.104.039156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Seimiya M., Kloter U., Flister S. and Gehring W. J. (1999). Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 126, 2253-2260. [DOI] [PubMed] [Google Scholar]
- Ohto H., Kamada S., Tago K., Tominaga S.-I., Ozaki H., Sato S. and Kawakami K. (1999). Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19, 6815-6824. 10.1128/MCB.19.10.6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. M., Rebay I., Tjian R. and Rubin G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137-147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Pappu K. S. (2003). Mechanism of hedgehog signaling during Drosophila eye development. Development 130, 3053-3062. 10.1242/dev.00534 [DOI] [PubMed] [Google Scholar]
- Pappu K. S., Ostrin E. J., Middlebrooks B. W., Sili B. T., Chen R., Atkins M. R., Gibbs R. and Mardon G. (2005). Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132, 2895-2905. 10.1242/dev.01869 [DOI] [PubMed] [Google Scholar]
- Peng H. W., Slattery M. and Mann R. S. (2009). Transcription factor choice in the Hippo signaling pathway: Homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 23, 2307-2319. 10.1101/gad.1820009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F., Hu B., Zavitz K. H., Xiao J., Garrity P. A. and Zipursky S. L. (1997). The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881-891. 10.1016/S0092-8674(00)80480-8 [DOI] [PubMed] [Google Scholar]
- Ray P., De S., Mitra A., Bezstarosti K., Demmers J. A. A., Pfeifer K. and Kassis J. A. (2016). Combgap contributes to recruitment of Polycomb group proteins in Drosophila. Proc. Natl. Acad. Sci. USA 113, 3826-3831. 10.1073/pnas.1520926113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready D. F., Hanson T. E. and Benzer S. (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217-240. 10.1016/0012-1606(76)90225-6 [DOI] [PubMed] [Google Scholar]
- Ruggiero R., Kale A., Thomas B. and Baker N. E. (2012). Mitosis in neurons: roughex and APC/C maintain cell cycle exit to prevent cytokinetic and axonal defects in drosophila photoreceptor neurons. PLoS Genet. 8, e1003049 10.1371/journal.pgen.1003049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer C. L. and Kumar J. P. (2009). Position dependent responses to discontinuities in the retinal determination network. Dev. Biol. 326, 121-130. 10.1016/j.ydbio.2008.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S. J., Davies E. L., Doyon L. and Rebay I. (2003). Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23, 5989-5999. 10.1128/MCB.23.17.5989-5999.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Chung S. and Kunes S. (2000). Combgap relays wingless signal reception to the determination of cortical cell fate in the Drosophila visual system. Mol. Cell 6, 1143-1154. 10.1016/S1097-2765(00)00112-X [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Clark M. J., Hoffmann F. M., Jackson P. D. and Brand H. (1994). Specificity of bone morphogenetic factors: cell fate and gene expression changes in drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 5, 585-593. [PubMed] [Google Scholar]
- Stowers R. S. and Schwarz T. L. (1999). A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152, 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova M. J. and Du W. (2008). Control of cell cycle entry and exiting from the second mitotic wave in the Drosophila developing eye. BMC Dev. Biol. 10, 1-10. https://doi.org/10.1186/1471-213x-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen P. C., Marshall S. D., Kyba M. and Brook W. J. (2000). The combgap locus encodes a zinc-finger protein that regulates cubitus interruptus during limb development in Drosophila melanogaster. Development 127, 4083-4093. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P. et al. (2015). STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447-D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T. and Mesirov J .P. (2013). Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178-192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrailas A. D. and Moses K. (2006). Smoothened, thickveins and the genetic control of cell cycle and cell fate in the developing Drosophila eye. Mech. Dev. 123, 151-165. 10.1016/j.mod.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Weasner B. M. and Kumar J. P. (2013). Competition among gene regulatory networks imposes order within the eye-antennal disc of Drosophila. Development 140, 205-215. 10.1242/dev.085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber J. L., Zhang J., Mitchell-Dick A. and Rebay I. (2013). 3D chromatin interactions organize Yan chromatin occupancy and repression at the even-skipped locus. Genes Dev. 27, 2293-2298. 10.1101/gad.225789.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T. and Ready D. F. (1991). The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 850, 841-850. [DOI] [PubMed] [Google Scholar]
- Xin S., Weng L., Xu J. and Du W. (2002). The role of RBF in developmentally regulated cell proliferation in the eye disc and in Cyclin D/Cdk4 induced cellular growth. Development 129, 1345-1356. [DOI] [PubMed] [Google Scholar]
- Xu P.-X., Cheng J., Epstein J. A. and Maas R. L. (1997). Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc. Natl. Acad. Sci. USA 94, 11974-11979. 10.1073/pnas.94.22.11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Canon J. and Banerjee U. (2003). A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev. Biol. 263, 323-329. 10.1016/j.ydbio.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Yang L. and Baker N. E. (2003). Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell 4, 359-369. 10.1016/S1534-5807(03)00059-5 [DOI] [PubMed] [Google Scholar]
- Yang L. and Baker N. E. (2006). Notch activity opposes ras-induced differentiation during the second mitotic wave of the developing Drosophila eye. BMC Cell Biol. 6, 8 10.1186/1471-213X-6-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Ranade S., Cai C. Q., Clouser C. and Pignoni F. (2006). Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881-4889. 10.1242/dev.02669 [DOI] [PubMed] [Google Scholar]
- Zhou Q., Zhang T., Jemc J. C., Chen Y., Chen R., Rebay I. and Pignoni F. (2014). Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contributions of Ey/Pax6 and So binding sites within two distant enhancers. Dev. Biol. 386, 152-164. 10.1016/j.ydbio.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. and Skoultchi A. I. (2001). Coordinating cell proliferation and differentiation. Curr. Opin. Genet. Dev. 11, 91-97. 10.1016/S0959-437X(00)00162-3 [DOI] [PubMed] [Google Scholar]