Abstract
The intrahepatic biliary network is a highly branched three-dimensional network lined by biliary epithelial cells, but how its branching patterns are precisely established is not clear. We designed a new computer-based algorithm that quantitatively computes the structural differences of the three-dimensional networks. Utilizing the algorithm, we showed that inhibition of Cyclin-dependent kinase 5 (Cdk5) led to reduced branching in the intrahepatic biliary network in zebrafish. Further, we identified a previously unappreciated downstream kinase cascade regulated by Cdk5. Pharmacological manipulations of this downstream kinase cascade produced a crowded branching defect in the intrahepatic biliary network and influenced actin dynamics in biliary epithelial cells. We generated larvae carrying a mutation in cdk5 regulatory subunit 1a (cdk5r1a), an essential activator of Cdk5. cdk5r1a mutant larvae show similar branching defects as those observed in Cdk5 inhibitor-treated larvae. A small-molecule compound that interferes with the downstream kinase cascade rescued the mutant phenotype. These results provide new insights into branching morphogenesis of the intrahepatic biliary network.
KEY WORDS: Biliary atresia, Intrahepatic biliary network, Actin dynamics, Biliary epithelial cells, Cdk5r1a
Highlighted article: Inactivation of Cdk5, or its regulator Cdk5r1a, leads to reduced network branching and disrupts actin organization in the zebrafish adult intrahepatic biliary network.
INTRODUCTION
The biliary system is responsible for transporting bile from the liver to the digestive tract. The intrahepatic biliary network, the biliary system within the liver, is a highly branched three-dimensional (3D) network that is pervasive throughout the liver. Biliary epithelial cells (also called cholangiocytes) are the innermost epithelial cells forming the conduit of this 3D network. Various means to visualize the 3D structure of the biliary network have been developed (Kaneko et al., 2015; Sparks et al., 2010). However, there is as yet no method to quantify differences in 3D branching patterns of the entire network, and this has presented a significant barrier to our understanding of the formation of the intrahepatic biliary network and the pathogenesis of liver diseases.
Cyclin-dependent kinase 5 (Cdk5) is a ubiquitously expressed serine-threonine kinase that is known to regulate a variety of biological processes, including cell migration, maturation, cell-cell connection and cytoskeletal organization in the nervous system (Dhavan and Tsai, 2001; Gilmore et al., 1998; Lagace et al., 2008; Ohshima et al., 1996). Unlike other members of the cyclin-dependent kinase family, Cdk5 is activated by the non-cyclin co-factor Cdk5 regulatory subunit 1 (Cdk5r1; also known as p35) (Chae et al., 1997) and is not involved in cell cycle regulation (Dhavan and Tsai, 2001; Lalioti et al., 2010). While Cdk5 has been intensively studied in the context of neural development, several studies have begun to hint at an important role outside the nervous system (Dhavan and Tsai, 2001; Na et al., 2015; Wei et al., 2005). However, the function of Cdk5 in liver development, particularly in biliary network development, has not been studied and there is no evidence yet linking Cdk5 to the pathology of biliary atresia.
Cdk5 is known to phosphorylate many substrate proteins, including those regulating cytoskeletal organization (Grant et al., 2001), such as p21-activated kinase 1 (Pak1) (Nikolic et al., 1998; Kichina et al., 2010). Direct substrates of Pak1 that regulate cytoskeletal organization include the LIM kinases (LimKs) (Edwards et al., 1999). LimKs, in turn, regulate Cofilin activity by phosphorylation of serine 3 (Arber et al., 1998). Cofilin proteins are actin-depolymerizing factors and important regulators of actin dynamics (Bamburg, 1999). To our knowledge, there is no study showing that Cdk5 can regulate the Pak1-LimK-Cofilin kinase cascade in vivo. Indeed, the roles of the Cdk5-mediated kinase cascade during biliary network formation have not been investigated.
Zebrafish has emerged as an excellent model to study human liver diseases (Goessling and Sadler, 2015). In zebrafish, the intrahepatic biliary network starts developing at 2 days post fertilization (dpf), before the biliary system starts to function at 5 dpf (Lorent et al., 2010; Sakaguchi et al., 2008). In mammals, the apical membranes of two adjacent hepatocytes generate the bile canaliculus, which secretes bile into the intrahepatic biliary network; however, in zebrafish the bile canaliculi are unicellular structures and tubular invaginations of the hepatocyte apical membranes (Lorent et al., 2004; Sakaguchi et al., 2008). In zebrafish, the intrahepatic biliary network is formed by small-diameter ductular biliary epithelial cells, which might be analogous to small-diameter ductular biliary epithelial cells in the mammalian liver lobule that transport bile from the canalicular network to the intralobular bile ducts in the portral tract (Kaneko et al., 2015). Indeed, there is remarkable conservation of both genes and gene function across vertebrate biliary system development, and thus many candidate genes responsible for biliary system abnormalities in humans can be screened efficiently and rapidly in the zebrafish model (Cui et al., 2011; Delous et al., 2012; Hand et al., 2009; Lorent et al., 2015, 2010, 2004; Matthews et al., 2011, 2005; Sakaguchi et al., 2008; Schaub et al., 2012). The zebrafish transgenic line Tg(Tp1-MmHbb:EGFP)um14, which is generated by conjugating tandem RBPJκ response element repeats with enhanced green fluorescent protein (EGFP), expresses EGFP specifically in the intrahepatic biliary network in the zebrafish liver (Lorent et al., 2010; Parsons et al., 2009). This transgenic line allowed us to visualize the entire intrahepatic biliary network in zebrafish larvae, greatly facilitating investigation of the development of the biliary system. Live imaging and other data from Lorent et al. (2010) with this transgenic line have clearly showed plasticity of intercellular connections in the developing biliary network. However, the study of the intrahepatic biliary network has been greatly inhibited by the lack of quantitative methods to measure the differences in aberrant branching patterns.
In this study, we developed a computational algorithm to quantify the differences in the branching patterns of the 3D network with a high degree of consistency and accuracy. Utilizing the algorithm, we identified a Cdk5-mediated kinase cascade that regulates branching and cellular actin organization of the intrahepatic biliary network. Finally, we provide genetic evidence that the kinase cascade functions in vivo to regulate proper branching of the intrahepatic biliary network.
RESULTS
Establishing a computational algorithm to quantify structural differences in 3D branching patterns
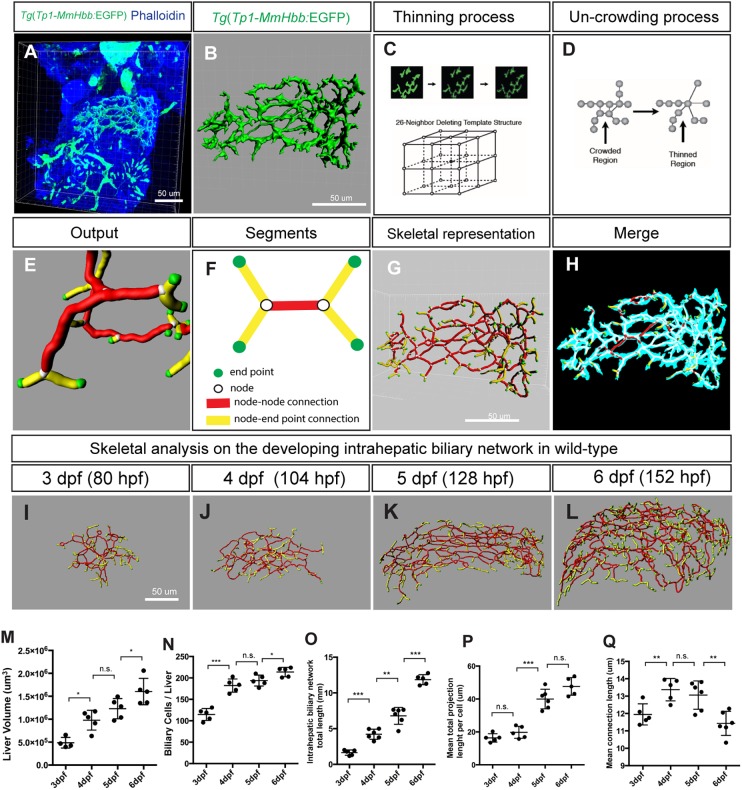
The intrahepatic biliary network is responsible for transporting bile from hepatocytes to the extrahepatic bile duct. This network possesses a highly branched and open-ended tree-like structure, which drains bile to a single exit that is connected to the extrahepatic duct. Subtle differences in the 3D branching of the intrahepatic biliary network can influence the function of the organ. In zebrafish, the Notch activity reporter line Tg(Tp1-MmHbb:EGFP)um14 (Lorent et al., 2010; Parsons et al., 2009) enabled us to visualize the entire intrahepatic biliary network (Fig. 1A,B). However, we lacked a method to consistently quantify differences in the branching patterns of 3D networks. To address this, we developed a computational algorithm capable of quantifying differences in network structures with a high degree of consistency and accuracy. The input data that we used for the computational algorithm were z-stack confocal scans of Tg(Tp1-MmHbb:EGFP)um14 expression in the entire liver (Fig. 1A). Since Tg(Tp1-MmHbb:EGFP)um14 is also expressed in tissues other than the liver, we digitally cropped the image such that only EGFP expression from the intrahepatic biliary network remained for analysis (Fig. 1B). Next, we adopted the previously published fully parallel 3D thinning algorithm (Ma and Sonka, 1996; Wang and Basu, 2007). This algorithm creates a compact representation of the intrahepatic biliary network marked by Tg(Tp1-MmHbb:EGFP)um14 expression, which we refer to as the ʻskeleton'. This algorithm repeatedly eliminates excess signals from EGFP-positive voxels in 3D space without changing the pattern of connectivity until the network is represented by continuous single voxels (Fig. 1C). Following this thinning process, Djikstra's shortest path algorithm (Cormen et al., 2009) was employed to thin any existing crowded regions, which generated a final unit-width skeleton (Fig. 1D). By combining these algorithms, the EGFP signals (Fig. 1B) are converted to skeletal representations of the network (Fig. 1E-G). As a result, the complex 3D network is represented by combinations of four basic segments: end point, node, node-node connection, and node-end point connection (Fig. 1E,F). To verify that our algorithm was working in the expected manner, we merged the original Tg(Tp1-MmHbb:EGFP)um14 expression data with the data that generated the skeleton of the network (Fig. 1H), and found that the two data sets completely overlapped. This suggests that the branching pattern analyses based on our algorithm are precise.
Fig. 1.
Skeletal analysis algorithm to quantify 3D branching patterns. (A) Projected confocal image of a Tg(Tp1-MmHbb:EGFP)um14 zebrafish liver visualized for EGFP and phalloidin expression at 4 dpf. (B) Segregated EGFP expression in the intrahepatic biliary network. (C) The thinning algorithm eliminates excess voxels from the surrounding 26 neighboring voxels until the network is presented by continuous single voxels. (D) The uncrowding algorithm eliminates congested areas, which are created by the thinning algorithm. (E) Graphical presentation of data generated from the skeletal analysis algorithm, which is the combination of the thinning and uncrowding algorithms. (F) Schematic representation of the skeletal analysis data. The complex 3D network is represented by a combination of four segments: end points, nodes, node-node connections, and node-end point connections. Each segment is given a unique identifier, and their location, length, shape, thickness and connectivity are defined. (G) Skeletal representation of the intrahepatic biliary network at 4 dpf generated by the skeletal analysis algorithm. (H) Merge of images in B and G. The original EGFP expression image and final skeletal representation match perfectly. (I-L) The skeletal analysis algorithm-based quantification of the developing intrahepatic biliary network. The skeletal representations of the intrahepatic biliary network were computed based on Tg(Tp1-MmHbb:EGFP)um14 expression in the wild-type liver at the indicated time points. (M) Average liver size (volume) of wild-type larvae. (N) Average number of Tg(Tp1-MmHbb:EGFP)um14-positive biliary epithelial cells in the liver. (O) The total length of the intrahepatic biliary network marked by Tg(Tp1-MmHbb:EGFP)um14 expression in the liver. (P) The total length of the intrahepatic biliary network divided by the number of biliary epithelial cells, indicating the mean projection length of the network per cell. (Q) Average length of connections in the liver. Error bars are s.d. *P<0.05, **P<0.01, ***P<0.001; n.s., not significant.
Algorithm-based analyses of intrahepatic biliary network formation in wild-type larvae
We first applied the skeletal analysis algorithm to quantify changes in the branching patterns of the intrahepatic biliary network at different time points during development in wild-type larvae (Fig. 1I-L, Table 1), analyzing Tg(Tp1-MmHbb:EGFP)um14 expression in the liver at 3, 4, 5 and 6 dpf. We measured biliary epithelial cell numbers by counting DAPI-stained nuclei of Tg(Tp1-MmHbb:EGFP)um14-expressing cells in the liver. Each parameter showed statistically significant differences between stages (Table 1), suggesting that this method could sensitively quantify differences in the branching patterns at these stages. We found that both liver size and total length of the intrahepatic biliary network increase linearly as larvae develop (Fig. 1M,O). Consistent with a previous study (Lorent et al., 2010), the number of Tg(Tp1-MmHbb:EGFP)um14-positive cells in the liver increased rapidly between 3 and 4 dpf, but then increased more slowly after 4 dpf (Fig. 1N). These results suggest that biliary epithelial cells proliferate before 4 dpf, and thereafter undergo remodeling to elongate. Consistent with this idea, we observed that before 4 dpf the nuclei of biliary epithelial cells cluster within the intrahepatic biliary network, but that these nuclei segregate by 5 dpf (Fig. S1), and the mean projection length of the intrahepatic biliary network per cell drastically increases between 4 and 5 dpf (Fig. 1P), suggesting that remodeling is taking place at this stage. Although both total network length and network length per cell increase, the average node-node connection length remains relatively constant from 3 to 6 dpf (Fig. 1Q), suggesting that new branches are always being generated during the elongation of the projections and connections. Overall, our algorithm-based method allowed us to measure the structural properties of the intrahepatic biliary network with a previously unachieved level of precision.
Table 1.
Structural parameters of the intrahepatic biliary network in wild-type larvae
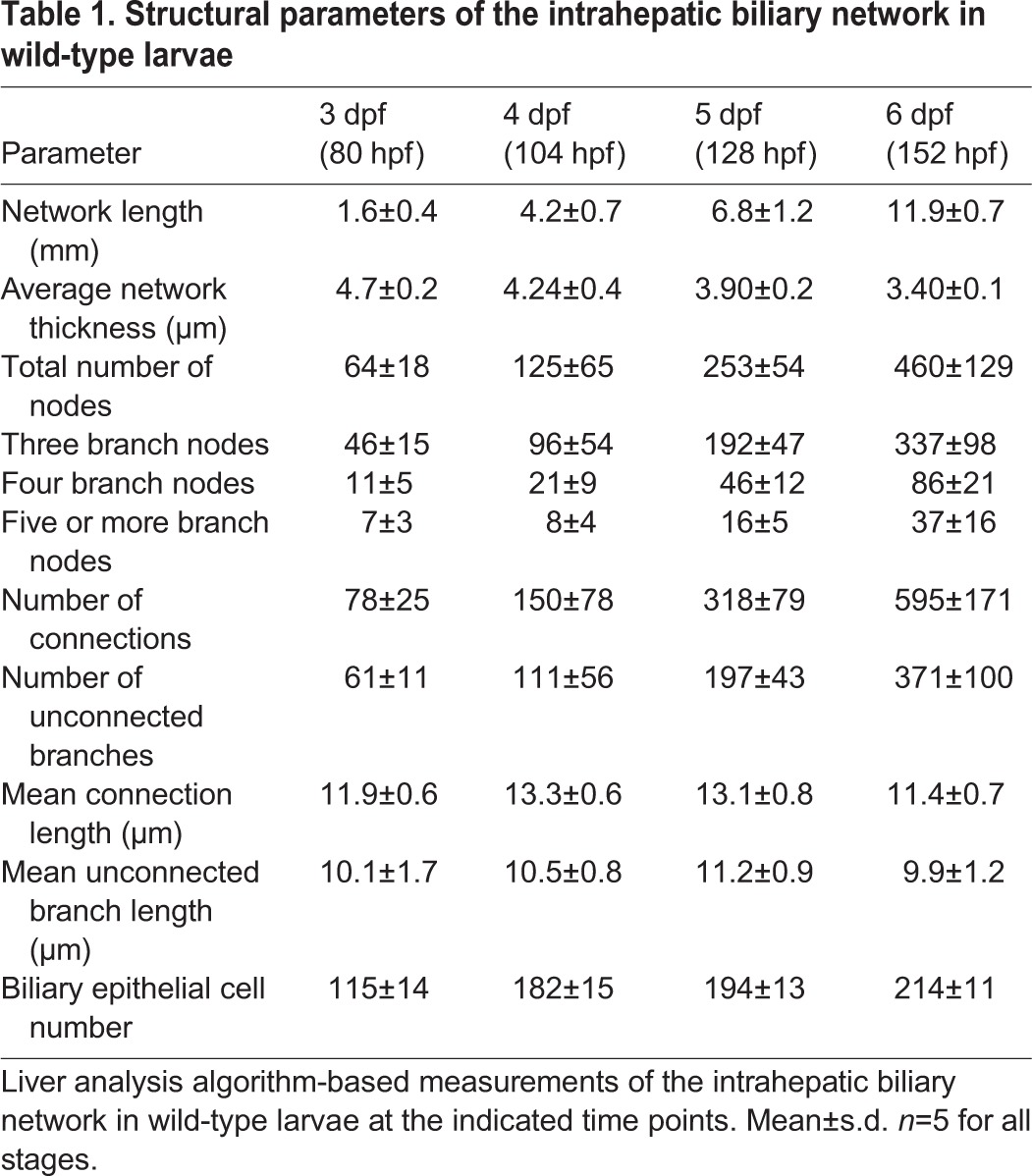
Cdk5 regulates intrahepatic biliary network formation
To investigate the molecular mechanisms underlying the formation of the intrahepatic biliary network, we used a small-molecule library to identify inhibitors affecting intrahepatic biliary network formation. We treated Tg(Tp1-MmHbb:EGFPum14/Tg(fabp10:RFP-CAAX)lri2 larvae with small-molecule compounds from 3 to 5 dpf and screened for those that significantly changed the branching pattern of the intrahepatic biliary network without affecting overall larval morphology, liver size or biliary epithelial cell numbers. We hypothesized that, with these screening criteria, we might be able to identify a new signaling pathway that specifically interfered with branching and remodeling processes without influencing cell differentiation. The only compound that produced a phenotype with these criteria was olomoucine (Sigma) (Wei et al., 2005). Compared with DMSO-treated controls, when Tg(Tp1-MmHbb:EGFP)um14 larvae were treated with 10 μM olomoucine from 3 to 5 dpf, they showed significantly altered branching patterns of the intrahepatic biliary network at 5 dpf (Fig. 2A,B) without obvious morphological changes to their physical appearance (Fig. S2). Further characterization of olomoucine-treated larvae with the skeletal analysis algorithm revealed that neither liver volume nor the number of biliary epithelial cells was significantly changed at 5 dpf (Fig. 2C,D), but that the mean total network length per cell was significantly reduced (Fig. 2E). The average number of connections per cell was also reduced (Fig. 2F), and the ratio of unconnected to connected branches increased in olomoucine-treated larvae (Fig. 2G). Olomoucine is a specific inhibitor of Cdk5, suggesting a novel role of Cdk5 in the development of the intrahepatic biliary network. These data suggest that, upon Cdk5 inhibition, biliary epithelial cells can properly differentiate and extend projections but that these projections cannot sufficiently elongate or form new connections.
Fig. 2.
Cdk5 regulates intrahepatic biliary network formation. (A,B) Skeletal representation of the intrahepatic biliary network in Tg(Tp1-MmHbb:EGFP)um14 larvae treated from 3 to 5 dpf with DMSO (A) or the Cdk5 inhibitor olomoucine (B) computed based on Tg(Tp1-MmHbb:EGFP)um14 expression at 5 dpf. Boxed regions in A and B are magnified in A′ and B′. Nodes, end points, connected (node-node) and unconnected (node-end point) branches are colored as in Fig. 1F. (C-G) Measurements of intrahepatic biliary network structures by the skeletal analysis algorithm of the DMSO-treated and olomoucine-treated Tg(Tp1-MmHbb:EGFP)um14 larvae at 5 dpf. (C) Mean liver volume at 5 dpf. Liver volume was not affected by olomoucine treatment. (D) Average number of Tg(Tp1-MmHbb:EGFP)um14-positive biliary epithelial cells in the liver at 5 dpf. (E) Mean total projection length of the intrahepatic biliary network normalized to biliary epithelial cell number. (F) Number of connected branches normalized to biliary epithelial cell number. (G) Ratio of connected to unconnected branches. n=5 for all measurements. (H,I) Projected images of confocal z-stacks of the liver in DMSO-treated (H) or olomoucine-treated (I) larvae visualized for the bile canaliculus marker Abcb11 (red) and Tg(Tp1-MmHbb:EGFP)um14 (green) expression at 5 dpf. (J) Average length of canaliculus measured based on Abcb11 expression. In total, 135 canaliculi were analyzed. (K) The number of canaliculi connected per 40 μm of the Tg(Tp1-MmHbb:EGFP)um14-expressing intrahepatic biliary network (n=10 for each condition). Error bars are s.d. *P<0.05, **P<0.01; n.s., not significant.
To examine the functionality of the biliary system in olomoucine-treated larvae we used the PED6 fluorescence assay (Farber et al., 2001), in which fluorescent metabolites generated in the intestine are transported through the biliary system to the gallbladder. In wild-type larvae treated with 10 μM olomoucine from 3 to 5 dpf, an average 18.5±2.2% (±s.d.; n=81 from three independent experiments) of larvae lacked PED6 fluorescence in the gallbladder at 5 dpf (Fig. S2). By contrast, in DMSO-treated control larvae, there were no larvae that completely lacked PED6 fluorescence in the gallbladder at 5 dpf (n=159 from three independent experiments; Fig. S2). These data suggest that inhibiting Cdk5 impaired the functionality of the biliary system.
Since reduced PED6 fluorescence in the gallbladder of olomoucine-treated larvae could be due to hepatic canalicular defects, we examined canaliculi with the Abcb11 antibody (Sakaguchi et al., 2008). Canaliculi did not appear to be influenced in olomoucine-treated larvae (Fig. 2H,I). Neither the average length of canaliculi (Fig. 2J) nor the frequency of canalicular connections to the intrahepatic biliary network (Fig. 2K) was changed in olomoucine-treated larvae, suggesting that reduced PED6 fluorescence in olomoucine-treated larvae is not due to defects in canaliculi. Consistently, in olomoucine-treated larvae we did not observe any changes in the expression of another biliary system marker, Cytokeratin (Ks18.4) (Fig. S3, Table S1) (Lorent et al., 2004; Matthews et al., 2005). These data suggest that olomoucine treatment influenced the intrahepatic biliary network without impacting the differentiation of biliary epithelial cells or the formation of canaliculi.
Cdk5 suppresses Pak1 activation in biliary epithelial cells
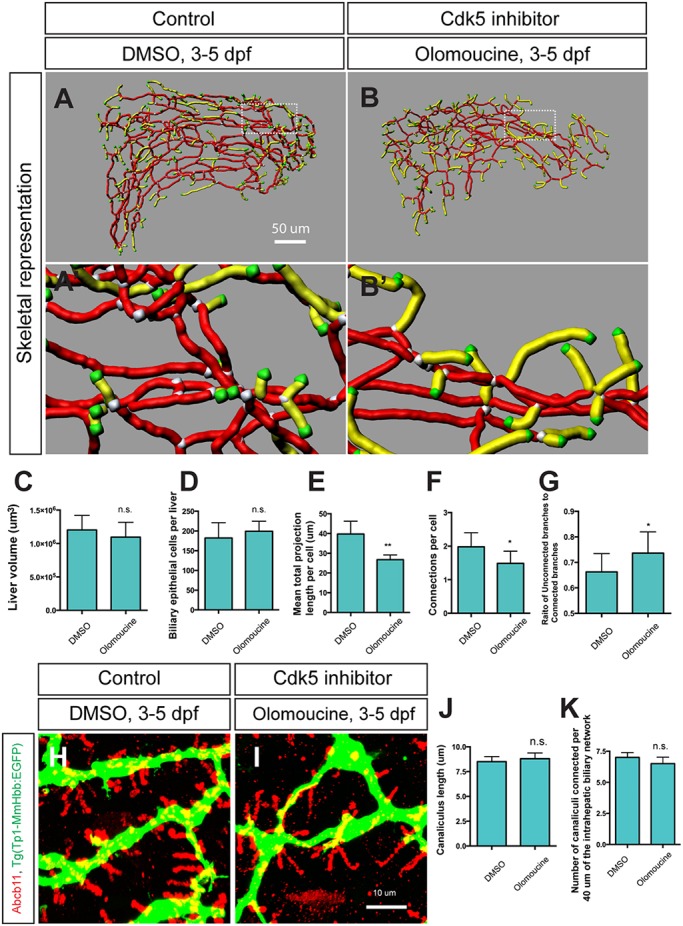
In order to understand the mechanisms by which Cdk5 regulates intrahepatic biliary network formation, we investigated downstream effectors regulated by Cdk5 in biliary epithelial cells. We tested small-molecule inhibitors against several known Cdk5 targets, and observed that Pak1 inhibitor treatment of wild-type larvae influenced the branching pattern of the intrahepatic biliary network. Pak1 is a direct target of Cdk5 and is known to regulate cytoskeletal dynamics (Nikolic et al., 1998). Based on this preliminary observation, we hypothesized that Pak1 might act downstream of Cdk5 to regulate intrahepatic biliary system formation. Cdk5 phosphorylates Pak1 on threonine 212 (T212) (Rashid et al., 2001), which inhibits Pak1 kinase activity. Pak1 activity can be measured by monitoring the activity-associated phosphorylation on threonine 423 (T423) (Zenke et al., 1999). Consistent with Cdk5 function in the nervous system, we found that inhibiting Cdk5 activated Pak1 in olomoucine-treated larvae at 5 dpf (Fig. 3A). In the DMSO-treated control liver, the baseline level of phospho-Pak1 (T423) exists predominantly in Tg(Tp1-MmHbb:EGFP)um14-expressing cells (Fig. 3B), suggesting that Pak1 is predominantly activated in biliary epithelial cells. In olomoucine-treated larvae, phospho-Pak1 (T423) levels increased predominantly in Tg(Tp1-MmHbb:EGFP)um14-expressing cells within the liver (Fig. 3C). These data suggest that suppressing Cdk5 function increases Pak1 activity primarily in biliary epithelial cells, which in turn suppresses branching morphogenesis.
Fig. 3.
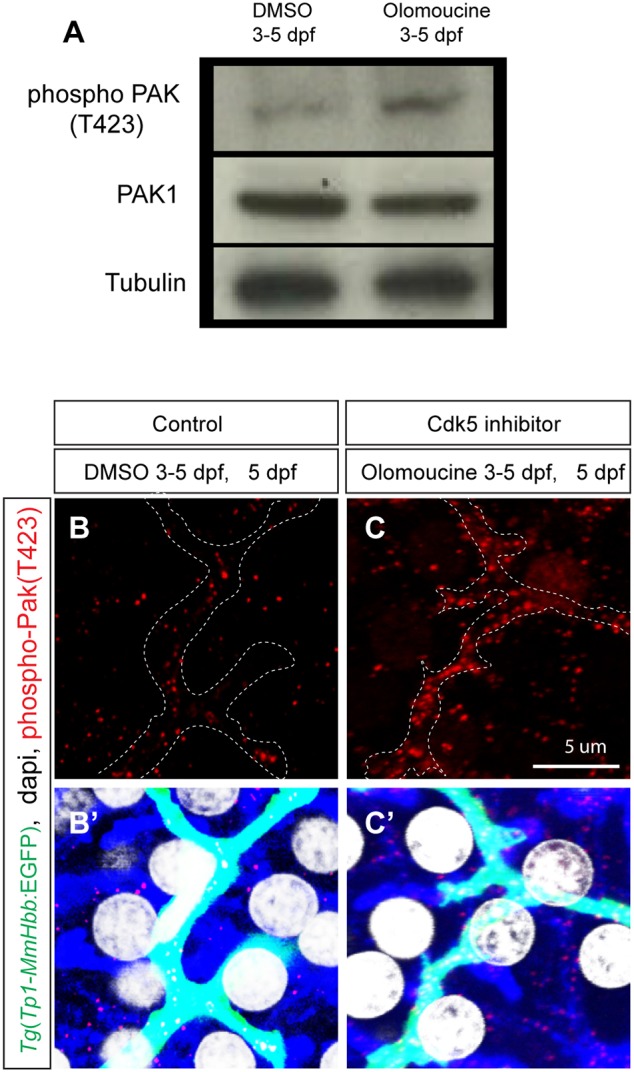
Cdk5 regulates Pak1 activity in biliary epithelial cells. (A) Pak1 phosphorylation (T423) levels in control DMSO-treated and olomoucine-treated wild-type Tg(Tp1-MmHbb:EGFP)um14 larvae were analyzed by western blotting at 5 dpf. Phospho-Pak1 was increased in olomoucine-treated larvae, whereas total Pak1 protein levels remained constant. Tubulin served as a loading control. These experiments were repeated three times with similar results. (B,C) Projected images of 4 μm-thick confocal z-stacks of the liver in DMSO-treated (B) or olomoucine-treated (C) larvae visualized for phospho-Pak1 (T423) (red). Biliary epithelial cells are outlined. (B′,C′) Phospho-Pak1 visualization merged with Tg(Tp1-MmHbb:EGFP)um14 expression (green), DAPI staining (white) and F-actin (phalloidin, blue). Pak1 is predominantly activated in Tg(Tp1-MmHbb:EGFP)um14-positive biliary epithelial cells in the liver of olomoucine-treated larvae.
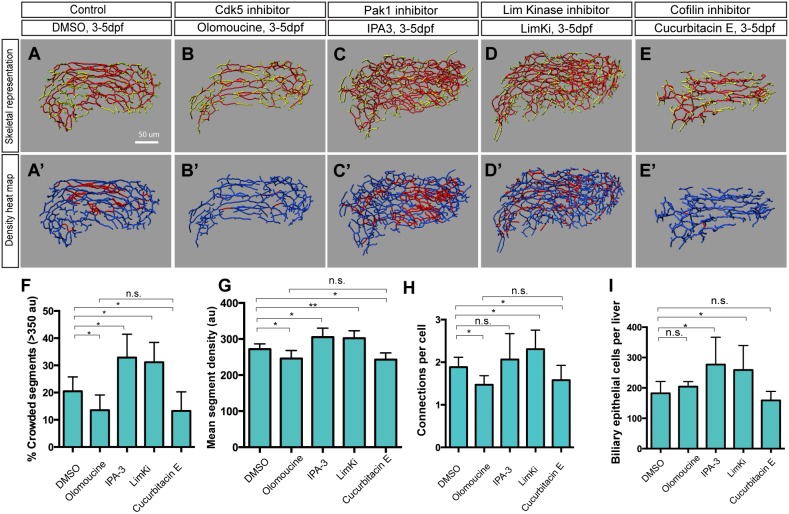
Consistent with this idea, treatment with 1 μM Pak1 inhibitor IPA-3 (Sigma) (Deacon et al., 2008) from 3-5 dpf generated an intrahepatic biliary network phenotype opposite to that observed in olomoucine-treated larvae; the density of the network appeared to be greater than that of DMSO-treated wild-type control larvae (Fig. 4A-C) without any observable change in larval physical appearance (Fig. S2C).
Fig. 4.
The Cdk5-Pak1-LimK-Cofilin kinase cascade regulates biliary system formation. (A-E) Skeletal representations of the intrahepatic biliary network in 5 dpf wild-type Tg(Tp1-MmHbb:EGFP)um14 larvae treated from 3 to 5 dpf with DMSO (A), olomoucine (B), IPA-3 (C), LimKi (D) or cucurbitacin E (E). (A′-E′) Density heat maps of the skeletal representations, in which segments with higher density [≥350 arbitrary units (au)] and lower density (<350 au) are colored red and blue, respectively. (F-I) Measurements of intrahepatic biliary network structures by the skeletal analysis algorithm of larvae treated as in A-E. (F) Percentage of segments showing high density (>350 au), which was calculated by the density analysis algorithm, as quantified from the density heat maps shown in A′-E′. (G) Mean segment density calculated by the density analysis algorithm. (H) Number of connected branches per biliary epithelial cell in the liver at 5 dpf. (I) Mean biliary epithelial cell number per liver. Error bars are s.d. n=5 for each treatment. *P<0.05, **P<0.01; n.s., not significant.
In order to quantify the density of the network, we calculated how many voxels existed within a sphere of 20 μm radius drawn on each segment (Fig. 4A′-C′). Both the percentage of higher density (>350 au) segments and the average segment density showed opposite trends in olomoucine-treated versus IPA-3-treated larvae at 5 dpf, i.e. both decreased in olomoucine-treated larvae and increased in IPA-3-treated larvae (Fig. 4F,G). It should be noted that in both olomoucine-treated and IPA-3-treated larvae, it appeared that the density of the intrahepatic biliary network was influenced uniformly throughout the entire network without any obvious regional differences within the liver (Fig. 4A′-C′), suggesting that suppressing Cdk5 or Pak1 influenced the intrahepatic biliary network without regional preference. These data together suggest that Pak1 activity in biliary epithelial cells is regulated by Cdk5 and is negatively correlated with the frequency of branch formation in the liver.
The Cdk5-Pak1-LimK-Cofilin kinase cascade regulates intrahepatic biliary network formation
We next investigated how the Cdk5-Pak1 kinase cascade regulates branching morphogenesis of the intrahepatic biliary network. In the nervous system, Pak1 is known to phosphorylate LimKs to regulate their activities (Edwards et al., 1999; Ray et al., 2014). LimKs, in turn, phosphorylate and thereby suppress the activity of Cofilin (Mokalled et al., 2010). We hypothesized that the Cdk5-Pak1-LimK-Cofilin kinase cascade might play a role in intrahepatic biliary network branching morphogenesis. We found that suppressing LimKs by treatment with 10 μM LimK inhibitor (LimKi; Millipore) (Ross-Macdonald et al., 2008) from 3-5 dpf generated a crowded intrahepatic biliary network phenotype similar to that observed in Pak1 inhibitor-treated larvae (Fig. 4D,F-I), without influencing larval physical appearance (Fig. S2). This supports the idea that the Pak1-LimK kinase cascade found in the nervous system is conserved in biliary epithelial cells. Consistently, suppressing Cofilin by treatment with 0.6 μM Cofilin inhibitor cucurbitacin E (Sigma) (Nakashima et al., 2010) from 3-5 dpf generated an intrahepatic biliary network phenotype similar to that observed in olomoucine-treated larvae (Fig. 4E-I). In IPA-3-treated and LimKi-treated larvae, the number of biliary epithelial cells was slightly increased (Fig. 4I). The number of biliary epithelial cells positive for mitosis-associated phospho-Histone H3 (Hans and Dimitrov, 2001) was increased in LimKi-treated larvae (Fig. S4), suggesting that suppressing LimK activity slightly increased biliary epithelial cell proliferation. Altogether, these data suggest that Cdk5 regulates branching morphogenesis of the intrahepatic biliary network through the Pak1-LimK-Cofilin kinase cascade.
The Cdk5-Pak1-LimK kinase cascade regulates Cofilin phosphorylation to regulate actin dynamics in biliary epithelial cells
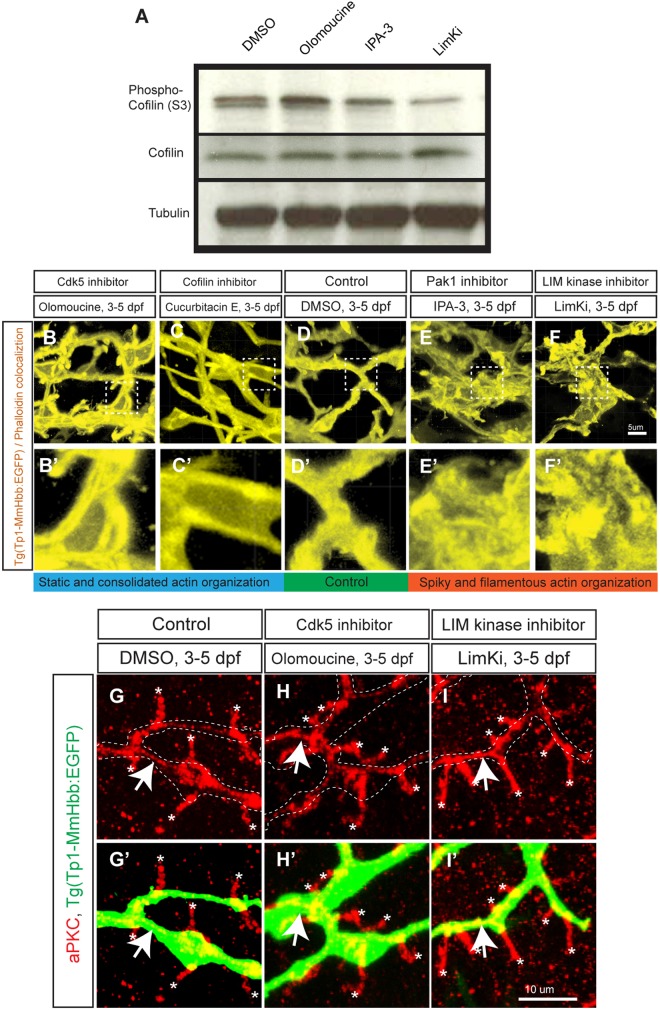
Based on these data, we hypothesize that Cdk5 inhibition increases Cofilin phosphorylation on serine 3 (S3) through the Pak1-LimK pathway, thereby suppressing the function of Cofilin. We examined the phosphorylation state of Cofilin S3 in larvae treated with olomoucine, IPA-3 or LimKi by western blotting (Fig. 5A), and found that Cofilin S3 phosphorylation was increased in olomoucine-treated and decreased in IPA-3-treated and LimKi-treated larvae at 5 dpf. These data suggest that Cdk5 activity is positively correlated with Cofilin activity.
Fig. 5.
The Cdk5-mediated kinase cascade regulates F-actin cytoskeletal organization. (A) Cofilin phosphorylation levels at serine 3 (S3) in wild-type larvae treated with DMSO, olomoucine, IPA-3 or LimKi and analyzed by western blotting. Olomoucine treatment increased Cofilin S3 phosphorylation, thus inhibiting its activity. IPA-3 and LimKi treatment suppressed Cofilin S3 phosphorylation, activating its activity. Total Cofilin protein levels remained constant. Tubulin blotting provided a loading control. These experiments were repeated three times with similar results. (B-F) Projected colocalization signal images computed based on Tg(Tp1-MmHbb:EGFP)um14 expression and phalloidin to visualize the actin network in biliary epithelial cells at 5 dpf in larvae treated as indicated. Compared with the control, F-actin appeared to be localized around the lumen in olomoucine-treated and cucurbitacin E-treated larvae, whereas it showed spiky and filamentous organization in IPA-3-treated and LimKi-treated larvae. Boxed regions in B-F are magnified in B′-F′. (G-I) Projected images of 15 μm-thick confocal z-stacks of the liver in larvae treated as indicated and visualized for apical membrane marker aPKC (red) expression at 5 dpf. (G′-I′) aPKC expression merged with Tg(Tp1-MmHbb:EGFP)um14 (green) expression. At 5 dpf, aPKC showed restricted expression in bile canaliculi (asterisks), which are the apical membranes of hepatocytes, and in the apical membranes of biliary epithelial cells (arrows) in control larvae. This expression was unchanged by olomoucine or LimKi treatment. These data suggest that Cdk5 or LimK inhibitors do not influence the apicobasal polarization of biliary epithelial cells.
Since Cofilin is an actin-severing factor that influences actin assembly and dynamics (Bravo-Cordero et al., 2013), we hypothesized that Cdk5 might regulate actin dynamics in biliary epithelial cells through the kinase cascade. To test this, we visualized the actin network in biliary epithelial cells in the liver by analyzing high-resolution confocal z-series scans in order to identify colocalization of signals between Tg(Tp1-MmHbb:EGFP)um14 and fluorescent phalloidin, a probe for filamentous actin (F-actin) (Fig. 5B-F). This visualization allowed us to observe the actin network in biliary epithelial cells with subcellular resolution in whole-mounted larvae. We found that in olomoucine-treated and cucurbitacin E-treated larvae, actin organization appeared to be consolidated as tubular structures within biliary epithelial cells (Fig. 5B,C), whereas in DMSO-treated control larvae the actin appeared to be localized relatively uniformly within the cell (Fig. 5D). In IPA-3-treated and LimKi-treated larvae, actin organization within biliary epithelial cells appeared spiky and filamentous (Fig. 5E,F). These data suggest that the Cdk5-Pak1-LimK-Cofilin kinase cascade regulates actin dynamics within biliary epithelial cells.
In order to assess whether modulated actin organization influences the polarization of biliary epithelial cells, we examined localization of the apical membrane marker aPKC (Sakaguchi et al., 2008) in olomoucine-treated and LimKi-treated larvae (Fig. 5G-I). We found no differences in aPKC localization within biliary epithelial cells, suggesting that, although it modulates actin dynamics, the Cdk5-Pak1-LimK-Cofilin kinase cascade does not influence the apicobasal polarization of biliary epithelial cells.
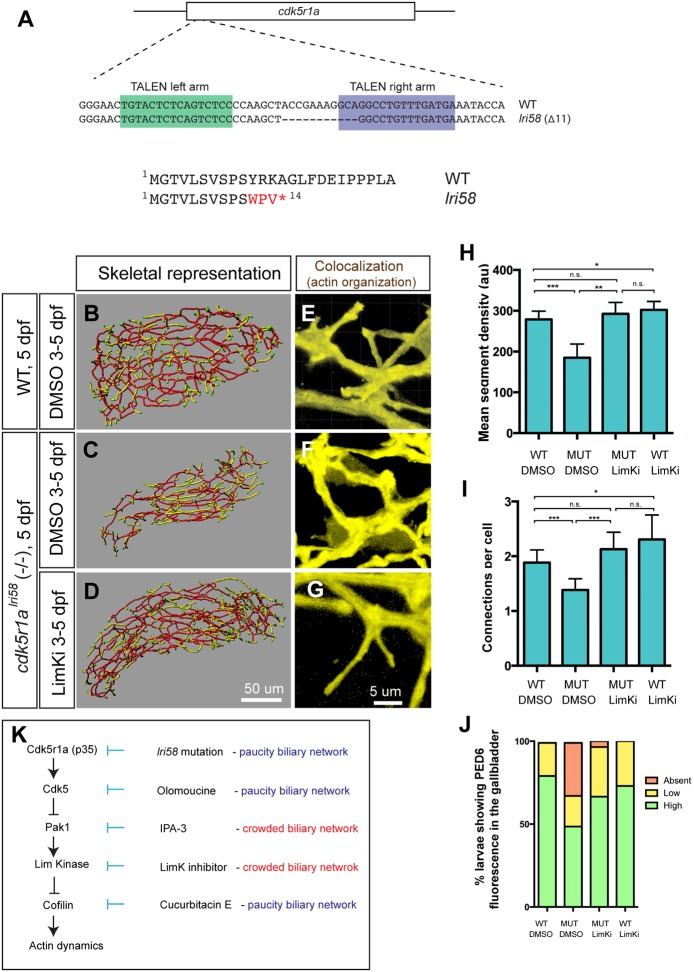
The intrahepatic biliary network phenotype of cdk5r1a knockout larvae can be rescued by manipulating the kinase cascade
In order to understand the upstream regulation of Cdk5 in biliary epithelial cells as well as to confirm that the kinase cascade we have identified utilizing small-molecule inhibitors actually functions in vivo, we generated cdk5r1a knockout zebrafish. We utilized Goldy TALENs targeting the first exon of the cdk5r1a gene. We then established the lri58 allele, in which 11 bp are deleted from the cdk5r1a gene, introducing a premature stop codon (Fig. 6A). cdk5r1alri58 mutant larvae occasionally develop small cardiac edema (Fig. S5) but otherwise their physical appearance remained normal at 5 dpf. However, Tg(Tp1-MmHbb:EGFP)um14 expression in the liver of cdk5r1alri58 larvae indicated that the intrahepatic biliary network appears to be less branched at 5 dpf (Fig. 6B,C), and further investigation based on the skeletal analysis algorithm confirmed that their mean segment density and number of connections are decreased compared with wild-type siblings (Fig. 6H,I). Furthermore, in cdk5r1alri58 larvae, actin organization in biliary epithelial cells is consolidated, as seen in larvae treated with Cdk5 or Cofilin inhibitors (compare Fig. 6F with Fig. 5B,F). These data suggest that Cdk5r1a functions in biliary epithelial cells to regulate Cdk5 activity, which in turn influences branching morphogenesis of the intrahepatic biliary network.
Fig. 6.
cdk5r1a (p35) mutant larvae show intrahepatic biliary network defects, which are rescued by LimKi treatment. (A) The TALEN-based cdk5r1a knockout strategy. Sequence-specific TALENs were designed against the first exon of the cdk5r1a gene, which includes the entire ORF. We established the lri58 allele in which 11 bp were deleted from the cdk5r1a gene, truncating the Cdk5r1a protein at the fourteenth amino acid. (B-D) Skeletal representations of the intrahepatic biliary network in DMSO-treated wild-type (B) and in DMSO-treated (C) or LimKi-treated (D) cdk5r1alri58 mutant larvae computed based on Tg(Tp1-MmHbb:EGFP)um14 expression in the liver at 5 dpf. (E-G) Projected colocalization signal images computed based on Tg(Tp1-MmHbb:EGFP)um14 expression and phalloidin to visualize the actin network in biliary epithelial cells of larvae treated as in B-D. (H) Mean segment density in DMSO-treated or LimKi-treated wild-type or cdk5r1alri58 mutant larvae at 5 dpf. (I) Number of connected branches per biliary epithelial cell in the liver of DMSO-treated or LimKi-treated wild-type or cdk5r1alri58 mutant larvae at 5 dpf. n=5 for all experiments. Error bars are s.d. *P<0.05, **P<0.01, ***P<0.001; n.s., not significant. (J) Percentage of larvae showing high, low and no PED6 fluorescence in the gallbladder of DMSO-treated or LimKi-treated wild-type or cdk5r1alri58 mutant larvae at 5 dpf. (K) Model of the Cdk5-mediated kinase cascade that regulates branching morphogenesis of the intrahepatic biliary network. Cdk5r1a appears to be responsible for Cdk5 activation in biliary epithelial cells. Cdk5 suppresses Pak1 activity, and thus suppressing Cdk5 by olomoucine enhances Pak1 activity. Suppressing Pak1 activity by IPA-3 generated a biliary network phenotype opposite to that of cdk5r1a mutant and olomoucine-treated larvae. Pak1 appears to regulate LimKs, and thus suppressing LimK activity with LimKi generated a similar biliary network phenotype to that of larvae in which Pak1 activity is suppressed. LimKs suppress Cofilin activity by direct phosphorylation. Suppressing Cofilin by cucurbitacin E thus generated a biliary phenotype opposite to that of larvae in which Pak1 or LimK activity was suppressed.
We hypothesized that if Cdk5r1a is regulating intrahepatic biliary network formation through the Cdk5-Pak1-LimK-Cofilin kinase cascade, then we could rescue its phenotype by manipulating the kinase cascade. Since the suppression of Cdk5 leads to hyperactivation of Pak1 and LimKs in biliary epithelial cells (Fig. 3), we reasoned that suppressing LimK activity should rescue the intrahepatic biliary network phenotype in cdk5r1alri58 larvae. We treated cdk5r1alri58 larvae with 10 μM LimKi from 3 to 5 dpf and found that they had an intrahepatic biliary network similar to that of wild-type control larvae (Fig. 6D). We confirmed that both the mean segment density and number of connections per cell were recovered to wild-type levels in LimKi-treated cdk5r1alri58 larvae at 5 dpf (Fig. 6H,I). These data suggest that inhibiting LimK activity rescues the intrahepatic biliary network phenotype of cdk5r1alri58 mutant larvae. We also observed that LimKi-treated cdk5r1alri58 larvae showed an actin organization phenotype similar to that of control larvae (Fig. 6G). Finally, to examine the functionality of the biliary system in cdk5r1alri58 larvae, we used the PED6 fluorescence assay. In cdk5r1alri58 larvae at 5 dpf, an average 32±2.8% (n=74 from three experiments) of larvae lacked PED6 fluorescence in the gallbladder (Fig. 6J, Fig. S2). However, in LimKi-treated cdk5r1alri58 larvae, the number showing no PED6 fluorescence in the gallbladder was greatly decreased (average 3.5±2.1%; n=80 from three experiments; P<0.01; Fig. 6J, Fig. S2). These data suggest that inhibiting LimK can rescue the functionality of the biliary system in cdk5r1alri58 mutant larvae, further supporting the hypothesis that the Cdk5-Pak1-LimK-Cofilin kinase cascade functions in vivo to regulate actin organization and branching patterns of biliary epithelial cells.
DISCUSSION
In this study, we developed a new quantitative tool to assess complex biliary network branching and connectivity. We utilized an innovative approach to establish computer-based structural analysis to quantify 3D branching differences with previously unachieved precision. By combining pharmacological and genetic manipulations with imaging technologies and image analysis algorithms, we have been able to quantify and analyze a variety of differences in 3D branching patterns that would be difficult to precisely measure by a less integrative and innovative approach.
For instance, we identified that olomoucine-treated larvae show reduced density in the intrahepatic biliary network, whereas liver size and biliary epithelial cell number remain constant in these larvae. Therefore, without using this algorithm, it would be challenging to analyze or quantify branching phenotypes of olomoucine-treated larvae, especially with conventional 2D sections of the liver. These facts further emphasize the importance of quantifying differences in 3D branching patterns. The limitation of this method is that rare Tg(Tp1-MmHbb:EGFP)um14-negative terminal cells will be excluded from this computational assay. The Anxa4 antibody (mAb 2F11) recognizes most of the biliary epithelial cells (Sakaguchi et al., 2008; Zhang et al., 2014), and this algorithm can also analyze the Anxa4 expression pattern (Fig. S6). It would also be possible to apply this algorithm, with minor modifications, to analyze branching patterns of other highly branched networks, such as vascular networks in other organs or organisms.
The Cdk5-Pak1-LimK-Cofilin kinase cascade regulates branching morphogenesis of the intrahepatic biliary network
In this study, we identified that the Cdk5-Pak1-LimK-Cofilin kinase cascade regulates branching morphogenesis of the intrahepatic biliary network (Fig. 6K). Previously in zebrafish, Pak1 was shown to be required for proper neural crest migration and cardiac outflow tract formation (Kelly et al., 2014). Consistent with this report, we found that IPA-3 treatment of wild-type embryos from 6 to 48 hours post fertilization (hpf) induced morphological and cardiac phenotypes (Fig. S2G), but treatment with IPA-3 from 3 to 5 dpf, as used in this study, did not affect blood circulation (Fig. S2C), suggesting that IPA-3 is inhibiting the function of Pak1 properly and that, after 3 dpf, Pak1 is not required for cardiac outflow tract formation but rather for proper intrahepatic biliary network formation.
Consistent with the fact that Cofilin is an actin-severing protein, we showed that manipulating the Cdk5-Pak1-LimK-Cofilin kinase cascade significantly impacted actin organization in biliary epithelial cells (Fig. 5B-F). We propose that this kinase cascade influences branching morphogenesis by regulating actin cytoskeletal dynamics in biliary epithelial cells. Suppressing Cdk5 function did not affect the number of biliary epithelial cells (Fig. 2D), indicating that biliary epithelial cell proliferation is not responsible for the difference in the intrahepatic biliary network; instead, Cdk5 suppression might reduce the availability of actin monomers required for elongating cellular projections of biliary epithelial cells to form new connections. Conversely, suppressing LimK functions might mobilize actin monomers to facilitate cellular elongation and the formation of new connections. Although suppressing LimK activity slightly increased biliary cell numbers by enhancing cell proliferation (Fig. 4I, Fig. S4), the number of connections per cell was still higher in LimKi-treated larvae, suggesting that biliary epithelial cells form more connections to increase the density of the network. This study does not address the role of the Cdk5-mediated kinase pathway in filopodia projection of biliary epithelial cells, and it would be interesting to investigate further the links between the Cdk5-mediated kinase cascade and previously observed filopodia projection patterns (Lorent et al., 2010) and planar cell polarity (Cui et al., 2011) in biliary epithelial cells. Since Cofilin is also known to regulate Myosin II binding to actin (Wiggan et al., 2012) and endocytosis (Okreglak and Drubin, 2007), we cannot rule out the possibility that Cofilin influences branching morphogenesis of the intrahepatic biliary network through a mechanism unrelated to actin dynamics.
The fact that we were able to rescue the biliary network phenotypes of cdk5r1alri58 larvae by treatment with LimKi to wild-type levels suggests that the kinase cascade that we have identified is a linear pathway responsible for most of the biliary phenotypes in these mutant larvae. Although Cdk5 is known to have many target substrates (Dhavan and Tsai, 2001), these results suggest that the Pak1-LimK-Cofilin kinase cascade is the major Cdk5 target in the biliary epithelial cells of larval zebrafish.
The pathogenesis of biliary atresia and roles of the Cdk5-mediated kinase cascade
Biliary atresia is a progressive obliterative disease in the biliary system that occurs exclusively in the embryonic and neonatal period (Mack and Sokol, 2005; Perlmutter and Shepherd, 2002; Sokol et al., 2003). Biliary atresia is not generally thought of as an inherited disease, although there are some lines of evidence to support the hypothesis that genetic factors contribute to disease susceptibility (Garcia-Barceló et al., 2010; Tang et al., 2016; Tsai et al., 2014). Our study has indicated that inhibiting the Cdk5-mediated pathway can generate phenotypes similar to those seen in biliary atresia patients. Although there is no direct evidence to indicate that the Cdk5-mediated pathway is involved in the pathogenesis of biliary atresia, there is a clinical report of impaired biliary system formation in a Williams syndrome patient (O'Reilly et al., 2006), in which the copy number of multiple genes, including LIMK1, is changed, suggesting that the kinase cascade pathway identified in the zebrafish model might also be functioning in humans. We found that treatment with LimKi can ameliorate the biliary network paucity phenotype as well as restore liver functionality in cdk5r1alri58 mutant larvae, suggesting that this small-molecule inhibitor might be useful to ameliorate liver functionality in cholangiopathy patients. It would be intriguing to test further the beneficial effects of LimKi on other zebrafish, as well as mammalian, biliary atresia models.
MATERIALS AND METHODS
Zebrafish husbandry and transgenic lines
Zebrafish (Danio rerio) larvae were obtained from crosses of the wild-type AB/TL strain or heterozygous mutant fish as previously described (Schaub et al., 2012). The following transgenic and mutant lines were used: Tg(fabp10:RFP-CAAX)lri2, Tg(Tp1-MmHbb:EGFP)um14 and cdk5r1alri58. We followed the protocol approved by the Cleveland Clinic's Institutional Animal Care and Use Committee.
Computational skeletal analyses with z-series confocal images
Confocal z-series data of Tg(Tp1-MmHbb:EGFP)um14 and Tg(fabp10:RFP-CAAX)lri2 expression were obtained using a Leica SP5 confocal microscope. We used Imaris 8.2 software (Bitplane) to digitally crop the image such that only EGFP expression from the intrahepatic biliary network remained for further analysis. Liver volume was calculated based on Tg(fabp10:RFP-CAAX)lri2 expression utilizing the Imaris software. A custom-developed software written in MatLab 2011b (MathWorks) and packaged as Liver Analysis Program 5.3 was used for all computational skeletal analyses. A description of the software design is provided in the supplementary Materials and Methods. Liver Analysis Program 5.3 software is available upon request. For all confocal images analyzed by Liver Analysis Program 5.3, we have confirmed the proper skeletal conversion by overlaying the original confocal image with the converted skeletal image as shown in Fig. 1H. This overlay confirmation is a critical step in using the software properly.
Pharmacological screening
One or two Tg(Tp1-MmHbb:EGFP)um14;Tg(fabp10:RFP-CAAX)lri2 larvae were transferred per well of a 96-well plate. Small-molecule compounds from the Screen-Well kinase inhibitor (Enzo Life Sciences) and the LOPAC®1280 (Sigma) libraries were added at final concentration 12.5 μM to the 96-well plates at 3 dpf. At 5 dpf, we screened for compounds that did not change the overt physical appearance of larvae or the liver size as marked by Tg(fabp10:RFP-CAAX)lri2 expression but influenced the intrahepatic biliary network branching pattern as marked by Tg(Tp1-MmHbb:EGFP)um14. We screened 1359 compounds. We then verified three compounds with at least five larvae and the only compound that was confirmed to change the branching pattern of the intrahepatic biliary network without changing biliary epithelial cell number in the liver, liver size nor overt physical appearance was olomoucine. We quantified the phenotype using the skeletal analysis algorithm.
Pharmacological treatments
The following treatments were used: olomoucine (Sigma, O0886; 10 μM), IPA-3 (Sigma, I2285; 1 μM), LIM kinase inhibitor (Millipore, 435930; 10 μM) and cucurbitacin E (Sigma, SML0577; 0.6 μM). All pharmacological treatments were administered from 80 to 128 hpf in 1% (v/v) dimethyl sulfoxide (DMSO).
Western blotting and immunohistochemistry
Western blotting and immunohistochemistry were performed as previously described (Lorent et al., 2004; Nussbaum et al., 2013) using the antibodies listed in Table S1.
Goldy TALEN constructions and genotyping of cdk5r1alri58
Goldy TALEN targeting the cdk5r1a gene was designed and constructed as previously published (Bedell et al., 2012). The cdk5r1a recognition sequences are: left TALEN, 5′-TGTACTCTCAGTCTCC-3′; right TALEN, 5′-TCATCAAACAGGCCTGC-3′. The following primers were used for genotyping the lri58 allele: forward, 5′-GATTTCGTATCAAAAGACCCTGTA-3′; reverse, 5′-GCCCGAGAGATTGGCACAAGACTGAG-3′. The lri58 mutation eliminated the StuI site from the PCR product amplified by these primers.
Statistics
All statistical tests were performed by unpaired, two-tailed t-test using Prism 6 for Mac OS X.
Acknowledgements
We thank Laura Nagy, Brian Perkins, Veronique Lefebvre and Oliver Wessely for critical reading of the manuscript; the LRI Molecular Screening Core for sharing small-molecule inhibitor libraries; and Olov Andersson for exchanging unpublished information on zebrafish cdk5 gene functions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.F.S.; Methodology: M.D., C.B., L.X.P., G.N., I.G., A.M.; Software: M.D., C.B., A.V., K.T., T.F.S.; Formal analysis: M.D., C.B., I.G., K.T.; Investigation: M.D., C.B., L.X.P., G.N., I.G., A.M.; Data curation: M.D., C.B., I.G., K.T.; Writing - original draft: C.B., L.X.P., T.F.S.; Writing - review & editing: L.X.P., T.F.S.; Visualization: M.D., C.B., I.G.; Supervision: T.F.S.; Funding acquisition: T.F.S.
Funding sources
This work was supported by grants from the National Institutes of Health (NIH) (R00 DK078138 and RO1 DK103637), Cleveland Digestive Diseases Research Core Center funded by the NIH (P30DK084576) and Lerner Research Institute Chair's Innovation Research Fund to T.F.S. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.147397.supplemental
References
- Arber S., Barbayannis F. A., Hanser H., Schneider C., Stanyon C. A., Bernard O. and Caroni P. (1998). Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 393, 805-809. 10.1038/31729 [DOI] [PubMed] [Google Scholar]
- Bamburg J. R. (1999). Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185-230. 10.1146/annurev.cellbio.15.1.185 [DOI] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., Krug R. G. III, Tan W., Penheiter S. G., Ma A. C., Leung A. Y. et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114-118. 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero J. J., Magalhaes M. A. O., Eddy R. J., Hodgson L. and Condeelis J. (2013). Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol. 14, 405-415. 10.1038/nrm3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae T., Kwon Y. T., Bronson R., Dikkes P., Li E. and Tsai L.-H. (1997). Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron 18, 29-42. 10.1016/S0896-6273(01)80044-1 [DOI] [PubMed] [Google Scholar]
- Cormen T. H., Leiserson C. E., Rivest R. L. and Stein C. (2009). Introduction to Algorithms, 3rd edn Cambridge, MA: MIT Press. [Google Scholar]
- Cui S., Capecci L. M. and Matthews R. P. (2011). Disruption of planar cell polarity activity leads to developmental biliary defects. Dev. Biol. 351, 229-241. 10.1016/j.ydbio.2010.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon S. W., Beeser A., Fukui J. A., Rennefahrt U. E. E., Myers C., Chernoff J. and Peterson J. R. (2008). An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem. Biol. 15, 322-331. 10.1016/j.chembiol.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delous M., Yin C., Shin D., Ninov N., Debrito Carten J., Pan L., Ma T. P., Farber S. A., Moens C. B. and Stainier D. Y. (2012). Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 8, e1002754 10.1371/journal.pgen.1002754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhavan R. and Tsai L.-H. (2001). A decade of CDK5. Nat. Rev. Mol. Cell Biol. 2, 749-759. 10.1038/35096019 [DOI] [PubMed] [Google Scholar]
- Edwards D. C., Sanders L. C., Bokoch G. M. and Gill G. N. (1999). Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253-259. 10.1038/12963 [DOI] [PubMed] [Google Scholar]
- Farber S. A., Pack M., Ho S. Y., Johnson I. D., Wagner D. S., Dosch R., Mullins M. C., Hendrickson H. S., Hendrickson E. K. and Halpern M. E. (2001). Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science 292, 1385-1388. 10.1126/science.1060418 [DOI] [PubMed] [Google Scholar]
- Garcia-Barceló M.-M., Yeung M.-Y., Miao X.-P., Tang C. S.-M., Cheng G., So M.-T., Ngan E. S.-W., Lui V. C.-H., Chen Y., Liu X.-L. et al. (2010). Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum. Mol. Genet. 19, 2917-2925. 10.1093/hmg/ddq196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore E. C., Ohshima T., Goffinet A. M., Kulkarni A. B. and Herrup K. (1998). Cyclin-dependent kinase 5-deficient mice demonstrate novel developmental arrest in cerebral cortex. J. Neurosci. 18, 6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W. and Sadler K. C. (2015). Zebrafish: an important tool for liver disease research. Gastroenterology 149, 1361-1377. 10.1053/j.gastro.2015.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P., Sharma P. and Pant H. C. (2001). Cyclin-dependent protein kinase 5 (Cdk5) and the regulation of neurofilament metabolism. Eur. J. Biochem. 268, 1534-1546. 10.1046/j.1432-1327.2001.02025.x [DOI] [PubMed] [Google Scholar]
- Hand N. J., Master Z. R., Eauclaire S. F., Weinblatt D. E., Matthews R. P. and Friedman J. R. (2009). The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology 136, 1081-1090. 10.1053/j.gastro.2008.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F. and Dimitrov S. (2001). Histone H3 phosphorylation and cell division. Oncogene 20, 3021-3027. 10.1038/sj.onc.1204326 [DOI] [PubMed] [Google Scholar]
- Kaneko K., Kamimoto K., Miyajima A. and Itoh T. (2015). Adaptive remodeling of the biliary architecture underlies liver homeostasis. Hepatology 61, 2056-2066. 10.1002/hep.27685 [DOI] [PubMed] [Google Scholar]
- Kelly M. L., Astsaturov A., Rhodes J. and Chernoff J. (2014). A Pak1/Erk signaling module acts through Gata6 to regulate cardiovascular development in zebrafish. Dev. Cell 29, 350-359. 10.1016/j.devcel.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichina J. V., Goc A., Al-Husein B., Somanath P. R. and Kandel E. S. (2010). PAK1 as a therapeutic target. Expert Opin. Ther. Targets 14, 703-725. 10.1517/14728222.2010.492779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace D. C., Benavides D. R., Kansy J. W., Mapelli M., Greengard P., Bibb J. A. and Eisch A. J. (2008). Cdk5 is essential for adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. USA 105, 18567-18571. 10.1073/pnas.0810137105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti V., Pulido D. and Sandoval I. V. (2010). Cdk5, the multifunctional surveyor. Cell Cycle 9, 284-311. 10.4161/cc.9.2.10466 [DOI] [PubMed] [Google Scholar]
- Lorent K., Yeo S. Y., Oda T., Chandrasekharappa S., Chitnis A., Matthews R. P. and Pack M. (2004). Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development 131, 5753-5766. 10.1242/dev.01411 [DOI] [PubMed] [Google Scholar]
- Lorent K., Moore J. C., Siekmann A. F., Lawson N. and Pack M. (2010). Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev. Dyn. 239, 855-864. 10.1002/dvdy.22220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K., Gong W., Koo K. A., Waisbourd-Zinman O., Karjoo S., Zhao X., Sealy I., Kettleborough R. N., Stemple D. L., Windsor P. A. et al. (2015). Identification of a plant isoflavonoid that causes biliary atresia. Sci. Transl. Med. 7, 286ra267 10.1126/scitranslmed.aaa1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C. M. and Sonka M. (1996). A fully parallel 3D thinning algorithm and its applications. Comput. Vis. Image Und. 64, 420-433. 10.1006/cviu.1996.0069 [DOI] [Google Scholar]
- Mack C. L. and Sokol R. J. (2005). Unraveling the pathogenesis and etiology of biliary atresia. Pediatr. Res. 57, 87R-94R. 10.1203/01.PDR.0000159569.57354.47 [DOI] [PubMed] [Google Scholar]
- Matthews R. P., Plumb-Rudewiez N., Lorent K., Gissen P., Johnson C. A., Lemaigre F. and Pack M. (2005). Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development 132, 5295-5306. 10.1242/dev.02140 [DOI] [PubMed] [Google Scholar]
- Matthews R. P., Eauclaire S. F., Mugnier M., Lorent K., Cui S., Ross M. M., Zhang Z., Russo P. and Pack M. (2011). DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology 53, 905-914. 10.1002/hep.24106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled M. H., Johnson A., Kim Y., Oh J. and Olson E. N. (2010). Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137, 2365-2374. 10.1242/dev.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Y. R., Jung D., Gu G. J., Jang A. R., Suh Y.-H. and Seok S. H. (2015). The early synthesis of p35 and activation of CDK5 in LPS-stimulated macrophages suppresses interleukin-10 production. Sci. Signal. 8, ra121 10.1126/scisignal.aab3156 [DOI] [PubMed] [Google Scholar]
- Nakashima S., Matsuda H., Kurume A., Oda Y., Nakamura S., Yamashita M. and Yoshikawa M. (2010). Cucurbitacin E as a new inhibitor of cofilin phosphorylation in human leukemia U937 cells. Bioorg. Med. Chem. Lett. 20, 2994-2997. 10.1016/j.bmcl.2010.02.062 [DOI] [PubMed] [Google Scholar]
- Nikolic M., Chou M. M., Lu W., Mayer B. J. and Tsai L.-H. (1998). The p35/Cdk5 kinase is a neuron-specific Rac effector that inhibits Pak1 activity. Nature 395, 194-198. 10.1038/26034 [DOI] [PubMed] [Google Scholar]
- Nussbaum J. M., Liu L. J., Hasan S. A., Schaub M., McClendon A., Stainier D. Y. R. and Sakaguchi T. F. (2013). Homeostatic generation of reactive oxygen species protects the zebrafish liver from steatosis. Hepatology 58, 1326-1338. 10.1002/hep.26551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima T., Ward J. M., Huh C. G., Longenecker G., Veeranna G., Pant H. C., Brady R. O., Martin L. J. and Kulkarni A. B. (1996). Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. USA 93, 11173-11178. 10.1073/pnas.93.20.11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V. and Drubin D. G. (2007). Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178, 1251-1264. 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly K., Ahmed S. F., Murday V. and McGrogan P. (2006). Biliary hypoplasia in Williams syndrome. Arch. Dis. Child. 91, 420-421. 10.1136/adc.2005.074740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. J., Pisharath H., Yusuff S., Moore J. C., Siekmann A. F., Lawson N. and Leach S. D. (2009). Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898-912. 10.1016/j.mod.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter D. H. and Shepherd R. W. (2002). Extrahepatic biliary atresia: a disease or a phenotype? Hepatology 35, 1297-1304. 10.1053/jhep.2002.34170 [DOI] [PubMed] [Google Scholar]
- Rashid T., Banerjee M. and Nikolic M. (2001). Phosphorylation of Pak1 by the p35/Cdk5 kinase affects neuronal morphology. J. Biol. Chem. 276, 49043-49052. 10.1074/jbc.M105599200 [DOI] [PubMed] [Google Scholar]
- Ray S., Fanti J. A., Macedo D. P. and Larsen M. (2014). LIM kinase regulation of cytoskeletal dynamics is required for salivary gland branching morphogenesis. Mol. Biol. Cell 25, 2393-2407. 10.1091/mbc.E14-02-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald P., de Silva H., Guo Q., Xiao H., Hung C.-Y., Penhallow B., Markwalder J., He L., Attar R. M., Lin T. A. et al. (2008). Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol. Cancer Ther. 7, 3490-3498. 10.1158/1535-7163.MCT-08-0826 [DOI] [PubMed] [Google Scholar]
- Sakaguchi T. F., Sadler K. C., Crosnier C. and Stainier D. Y. R. (2008). Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr. Biol. 18, 1565-1571. 10.1016/j.cub.2008.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub M., Nussbaum J., Verkade H., Ober E. A., Stainier D. Y. R. and Sakaguchi T. F. (2012). Mutation of zebrafish Snapc4 is associated with loss of the intrahepatic biliary network. Dev. Biol. 363, 128-137. 10.1016/j.ydbio.2011.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol R. J., Mack C., Narkewicz M. R. and Karrer F. M. (2003). Pathogenesis and outcome of biliary atresia: current concepts. J. Pediatr. Gastroenterol. Nutr. 37, 4-21. 10.1097/00005176-200307000-00003 [DOI] [PubMed] [Google Scholar]
- Sparks E. E., Huppert K. A., Brown M. A., Washington M. K. and Huppert S. S. (2010). Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology 51, 1391-1400. 10.1002/hep.23431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang V., Cofer Z. C., Cui S., Sapp V., Loomes K. M. and Matthews R. P. (2016). Loss of a candidate biliary atresia susceptibility gene, add3a, causes biliary developmental defects in zebrafish. J. Pediatr. Gastroenterol. Nutr. 63, 524-530. 10.1097/MPG.0000000000001375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai E. A., Grochowski C. M., Loomes K. M., Bessho K., Hakonarson H., Bezerra J. A., Russo P. A., Haber B. A., Spinner N. B. and Devoto M. (2014). Replication of a GWAS signal in a Caucasian population implicates ADD3 in susceptibility to biliary atresia. Hum. Genet. 133, 235-243. 10.1007/s00439-013-1368-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. and Basu A. (2007). A note on ‘A fully parallel 3D thinning algorithm and its applications’. Pattern Recogn. Lett. 28, 501-506. 10.1016/j.patrec.2006.09.004 [DOI] [Google Scholar]
- Wei F.-Y., Nagashima K., Ohshima T., Saheki Y., Lu Y.-F., Matsushita M., Yamada Y., Mikoshiba K., Seino Y., Matsui H. et al. (2005). Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat. Med. 11, 1104-1108. 10.1038/nm1299 [DOI] [PubMed] [Google Scholar]
- Wiggan O., Shaw A. E., DeLuca J. G. and Bamburg J. R. (2012). ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Dev. Cell 22, 530-543. 10.1016/j.devcel.2011.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke F. T., King C. C., Bohl B. P. and Bokoch G. M. (1999). Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J. Biol. Chem. 274, 32565-32573. 10.1074/jbc.274.46.32565 [DOI] [PubMed] [Google Scholar]
- Zhang D., Golubkov V. S., Han W., Correa R. G., Zhou Y., Lee S., Strongin A. Y. and Dong P. D. S. (2014). Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev. Biol. 395, 96-110. 10.1016/j.ydbio.2014.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]