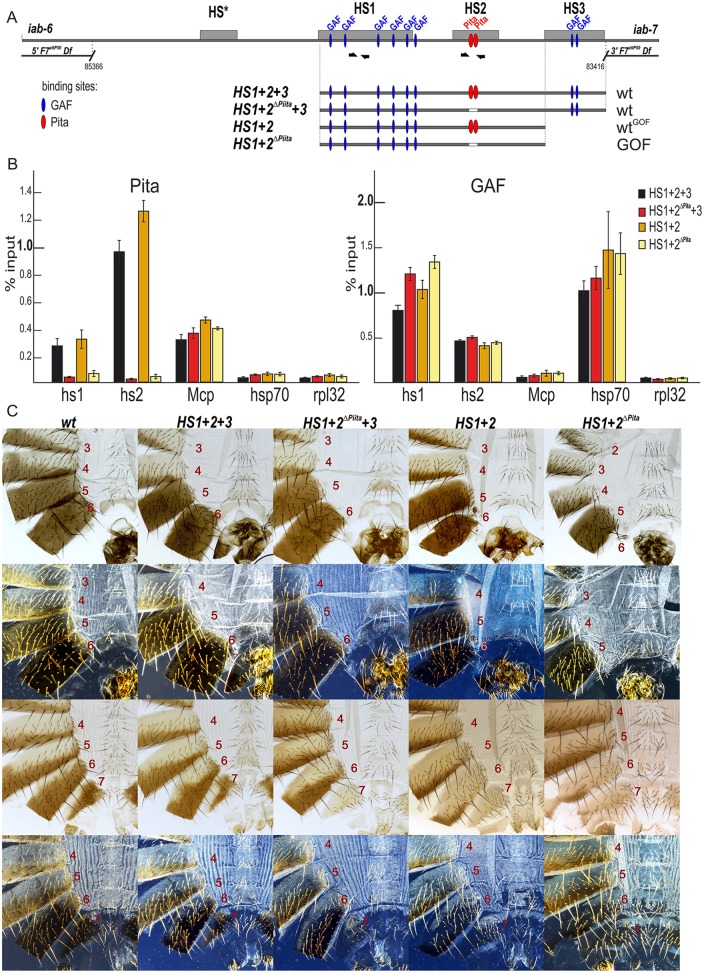
Fig. 3.
Role of Pita-binding sites in Fab-7 boundary function. (A) A schematic of the Fab-7 boundary replacement constructs at the F7attP50 site. Hypersensitive sites are shown as gray boxes. The proximal and distal deficiency endpoints of the F7attP50 deletions are shown. GAF- and Pita-binding sites are designated as blue and red ovals, respectively. Bold black half-arrows (marked i7) indicate primers that were used for ChIP experiments. (B) Binding of Pita and GAF at the HS1 and HS2 regions in pupae of tested transgenic lines (HS1+2+3, HS1+2ΔPita +3, HS1+2 and HS1+2ΔPita). The results of ChIP are presented as a percentage of the input DNA. The Mcp region was used as a positive control for Pita binding, the Hsp70 region was used as a positive control for GAF binding and the RpL32-coding region was used as a negative control for binding. Error bars indicate standard deviations of triplicate PCR measurements from two independent biological samples of chromatin. (C) All abdominal segments in HS1+2+3 and HS1+2ΔPita +3 males (first row, bright field; second row, dark field) and females (third row, bright field; fourth row, dark-field images) have essentially a wild-type identity. HS1+2 males have a mixed wild-type and gain-of-function phenotype: an almost complete A6 tergite in combination with an absence of the A6 sternite. HS1+2ΔPita males and females have a strong gain-of-function phenotype with small spots of cells with loss-of-function features.