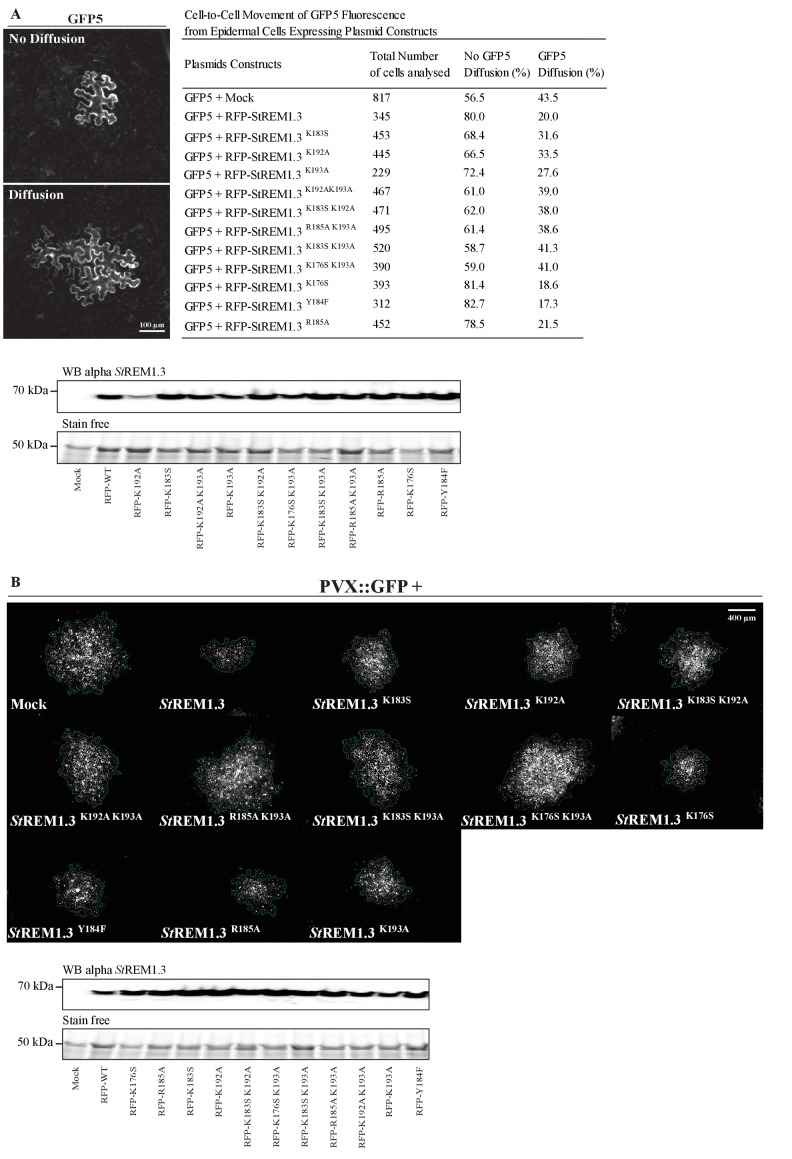
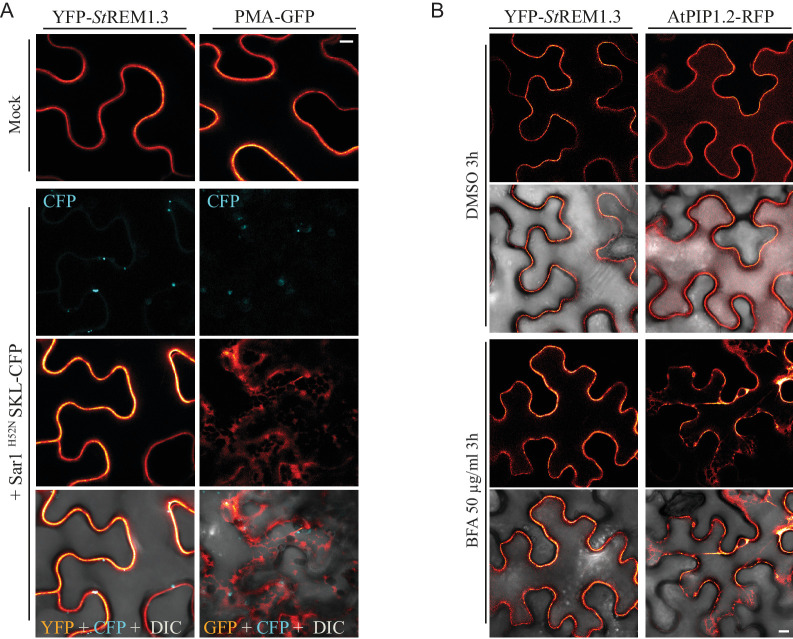
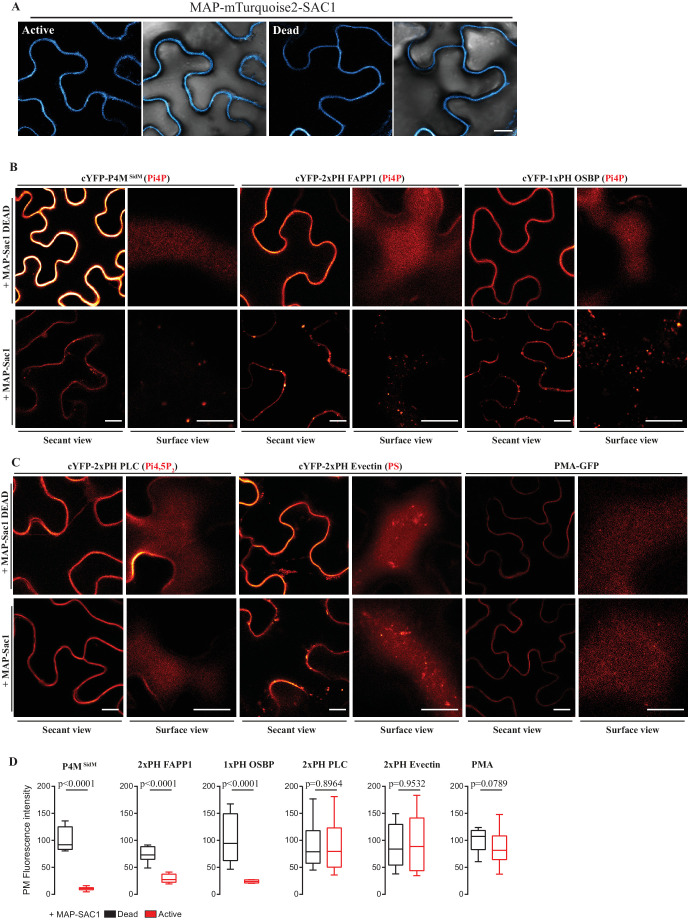
Figure 1. REMORIN localization into highly ordered PM nanodomains is mediated by sterols and PI4P.
( A ) Explanatory schematic of the secant or surface views of N. benthamiana leaf abaxial epidermal cell plasma membrane ( PM ) used throughout the article. ( B ) Confocal imaging surface views of Nicotinana benthamiana leaf epidermal cells expressing YFP- St REM1.3 with or without dominant-negative SAR1 H52N (PMA4-GFP was used as a potency control, see Figure 1—figure supplement 2 ), 24 hr after agroinfiltration. Tukey boxplots show the mean fluorescence intensity and the Spatial Clustering Index , SCI ( n = 3, quantification made on a representative experiment, at least 38 cells per condition). ( C ) Surface view confocal images showing the effect of Fenpropimorph (Fen) on PM patterning of YFP- St REM1.3 domains 20 hr after agroinfiltration. Tukey boxplots show the mean fluorescence intensity and the SCI of YFP- St REM1.3 in the Mock ( DMSO ) or Fen-treated leaves ( 50 µg / mL ), at least 46 cells from three independent experiments. ( D ) Secant view confocal fluorescence microscopy images displaying the degree of order of CFP- StREM1.3 ‐ enriched domains (left panel) by the environment ‐ sensitive probe Di ‐ 4 ‐ ANEPPDHQ (middle panel) 48 hr after agroinfiltration. Di ‐ 4 ‐ ANEPPDHQ red / green ratio ( RGM ) was measured for the global PM, and for the 10, 5, 2 % most intense CFP- St REM1.3 signal-associated pixels (right panel). A lower red / green ratio is associated with an increase in the global level of membrane order, at least 70 cells from three independent experiments. ( E ) Surface view confocal images showing the effect of dead or active constructs of MAP-SAC1p (MAP-mTurquoise2-SAC1p from yeast, see Figure 1—figure supplement 5 ) on PM domain localization of YFP- St REM1.3 20 hr after agroinfiltration. Tukey boxplots show the mean fluorescence intensity and the SCI of YFP- St REM1.3, at least 52 cells from four independent experiments. ( F ) Model showing the PI4P-driven targeting of the trimer of St REM1.3 to the PM and its PI4P- and sterol-dependent nanodomains organization. In all panels, p - values were determined by a two-tailed Mann-Whitney test.