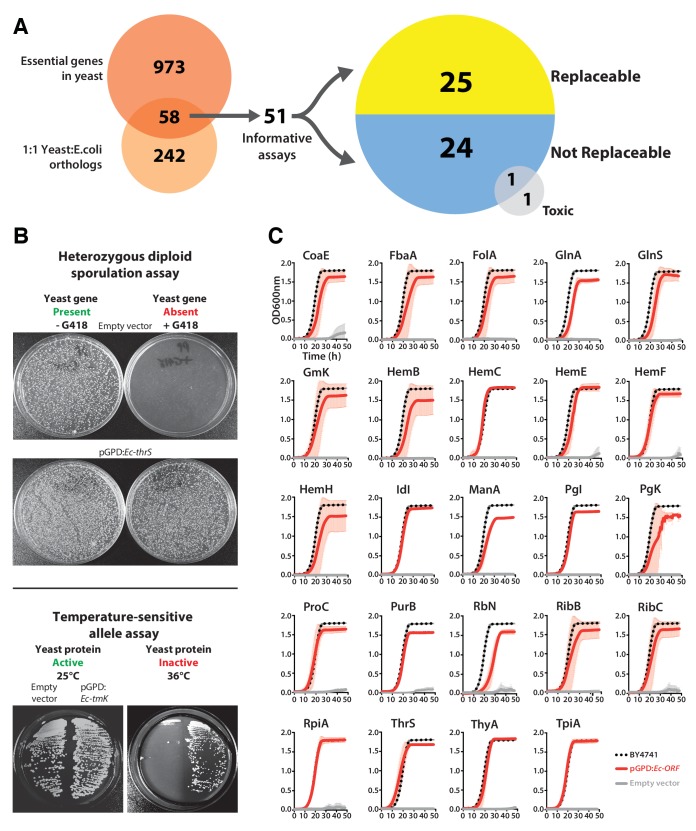
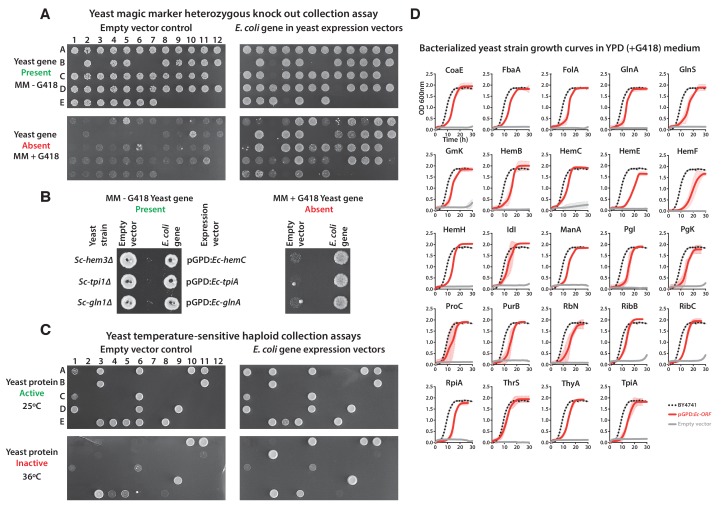
Figure 1. Many E. coli genes efficiently complement lethal growth defects in their yeast counterparts.
(A) Yeast and E. coli share hundreds of genes, 58 of which are essential in yeast and have clear 1:1 orthologs in either species. E. coli genes were cloned into a yeast expression vector under the control of a GPD promoter. 51 of these 58 E. coli genes provided informative assays for replaceability in yeast. Initial results from these complementation assays revealed that 25 of 51 (~49%) E. coli genes could functionally replace their orthologous yeast counterparts. (B) Complementation assays were performed in two different yeast strain backgrounds, as shown for representative assays. In the case of a yeast strain with a temperature-sensitive allele of the yeast gene Sc-cdc8, cells carrying the empty vector control grow at the permissive-temperature (25°C, yeast protein active) but not the restrictive-temperature (36°C, yeast protein inactive), unlike cells expressing the E. coli ortholog (Ec-tmK), indicating that the E. coli gene can functionally replace the yeast gene. In the case of yeast heterozygous diploid (Sc-ths1Δ/Sc-THS1) deletion strain, cells are sporulated and haploid progeny grown on selective medium (-Ura -Arg -His -Leu + Can) in the absence (yeast gene present) or presence of G418 (200 μg/ml) (yeast gene absent). Cells expressing the E. coli ortholog (Ec-thrS) grow on G418-containing medium, unlike cells carrying the empty vector control, indicating successful complementation. (C) Haploid yeast gene deletion strains carrying plasmids expressing functionally replacing E. coli genes (red solid-lines) generally exhibit comparable growth rates to the wild type parental yeast strain BY4741 (black dotted-lines). The empty vector control (grey solid-line) showed no such growth rescue in the presence of G418. Mean and standard deviation plotted with N = 3.