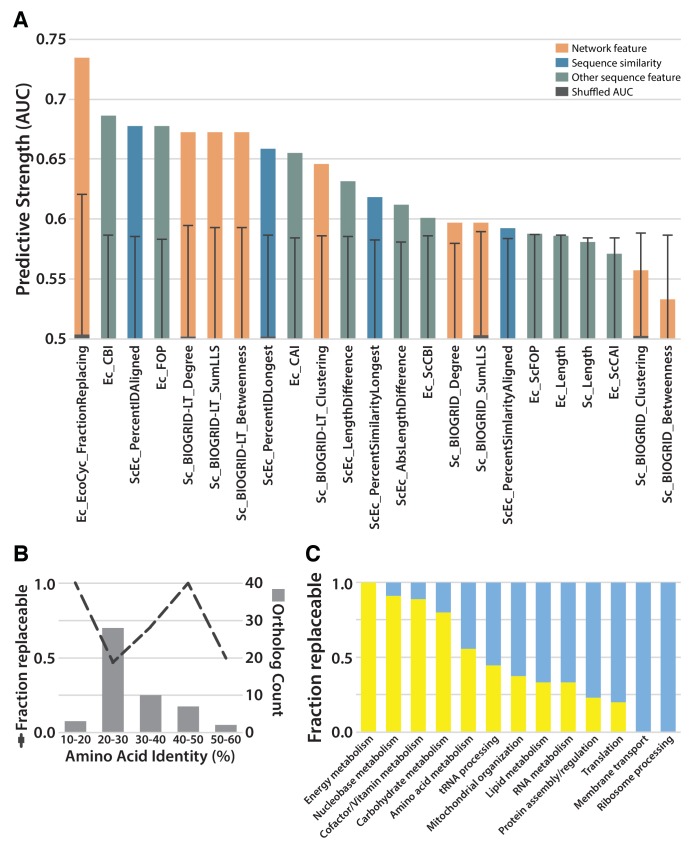
Figure 3. Replaceability of E. coli genes is a modular phenomenon.
(A) Several quantitative properties of the tested genes were assessed for their ability to predict replaceability, measured as the area under a receiver operating characteristic curve (AUC). Having a high fraction of interaction partners that replace was the most predictive property tested, suggesting that the ability to replace is a modular phenomenon whereby genes functioning together are similarly able to replace. A Random Forest classifier constructed with all attributes boosted the maximum AUC to 0.79. (B) As shown in (A), sequence similarity was not the most predictive feature. The fraction of replaceable genes in given ranges of similarity was variable, with the vast majority of orthologs being 20–30% identical, a range in which roughly half of proteins replaced. (C) Mapping of replaceability status onto yeast GO slim annotations revealed that GO categories have varying rates of replaceability, with core metabolic processes (e.g. energy metabolism, nucleobase metabolism) being largely replaceable while more specialized processes (e.g. protein assembly, membrane transport) were less so.