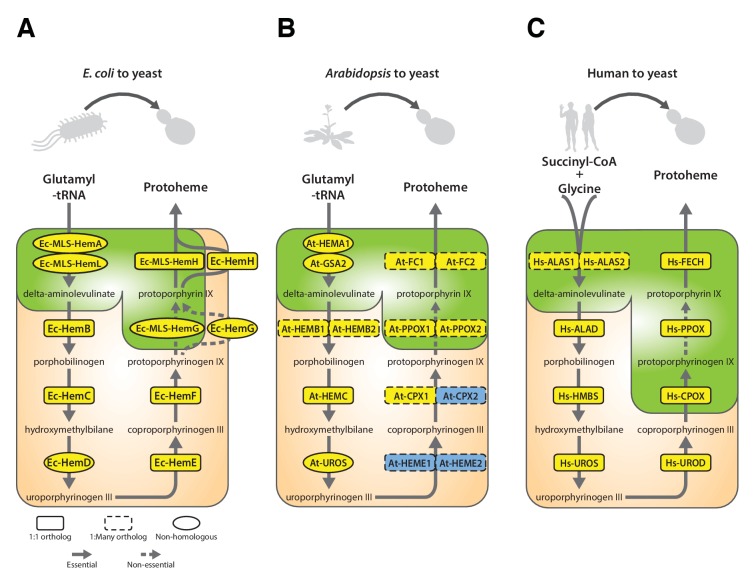
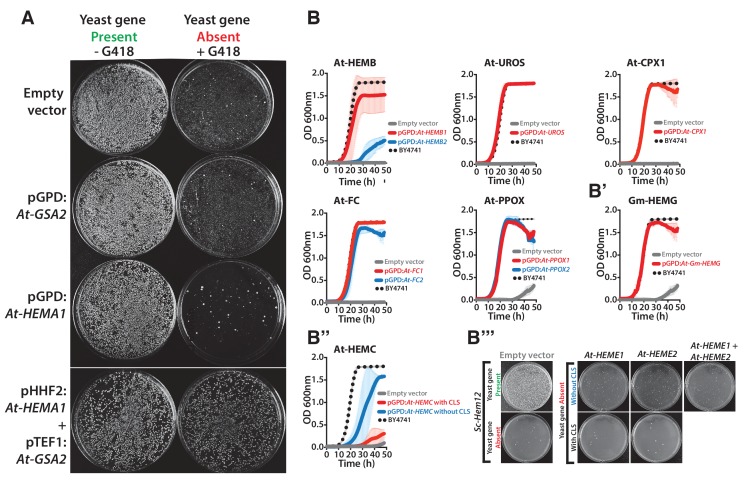
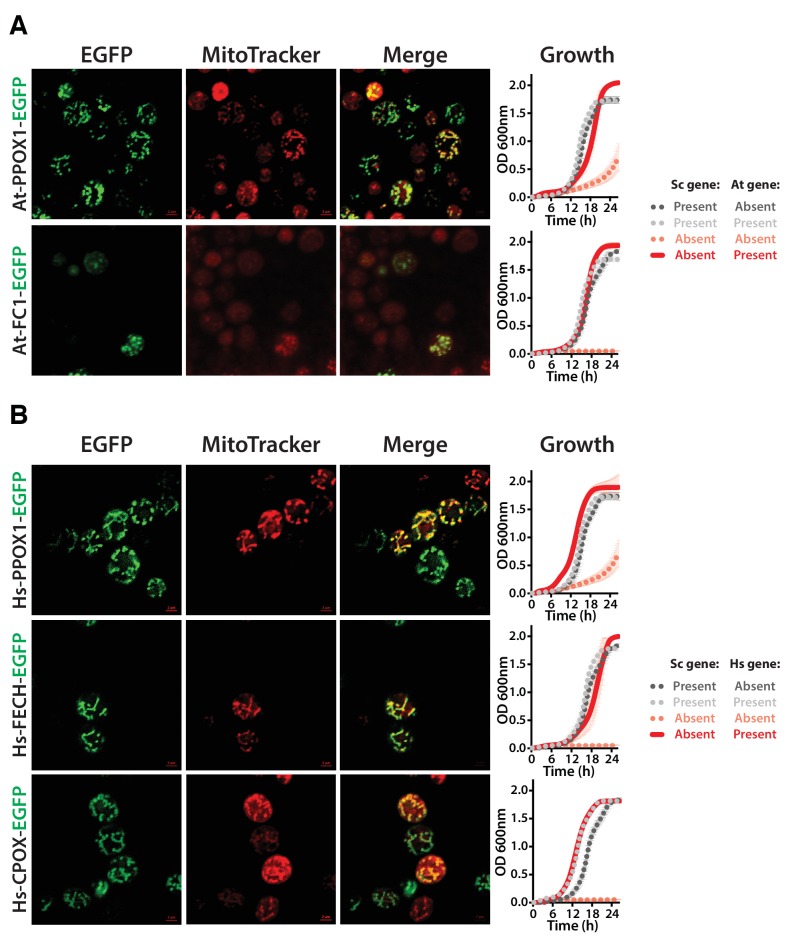
Figure 6. Yeast heme biosynthesis pathway enzymes can be successfully replaced by orthologs or analogs from bacteria, plants, and humans, in spite of alterations to subcellular localization.
Enzymatic steps of extant bacterial and eukaryotic heme biosynthesis pathways are identical save for the starting metabolites and conversion to delta-aminolevulinate; bacteria also exhibit non-orthologous gene displacement of several enzymes. Heme biosynthesis occurs in the bacterial cytoplasm and inner membrane, the human and yeast in mitochondria and cytoplasm, and the plant in chloroplast and cytoplasm. In spite of these localization changes over evolution, most of the defects in growth rate and viability conferred by heme pathway mutations in yeast can be complemented by introduction of the corresponding (A) bacterial genes, (B) plant genes (except for At-HemE), and (C) human genes. Yellow indicates a replaceable gene, blue non-replaceable.