Figure 1. Live-cell, 6-colour confocal microscopy to characterize the organelle interactome.
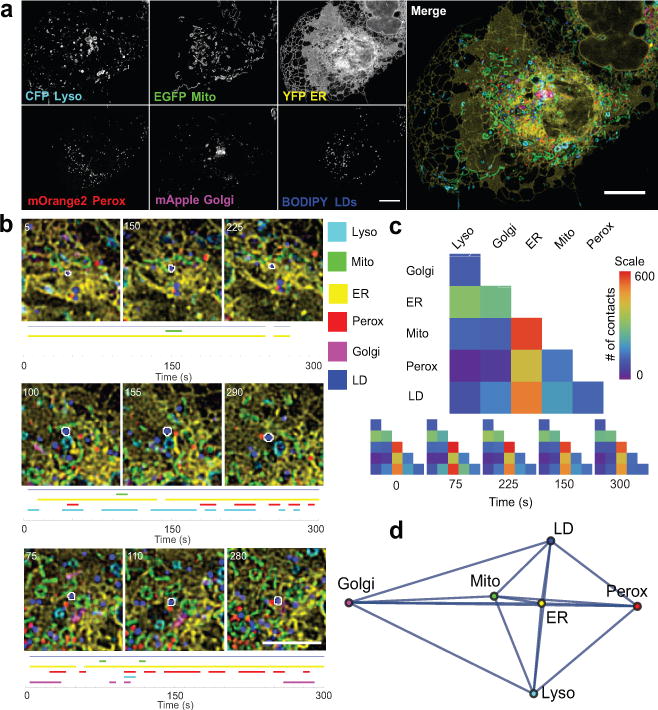
(a) Micrographs of a COS-7 cell expressing LAMP1-CFP, mito-EGFP, ss-YFP-KDEL, mOrange2-SKL, and mApple-SiT, and labelled with BODIPY 665/676. Micrographs are representative of 10 cells captured. (b) LDs (outlined in white) were tracked, and their interorganelle contacts mapped. A blue line indicates that the LD was successfully tracked, while coloured lines indicate that the LD was within 1 pixel (97 nm) of the indicated organelle at the specified time point. Numbers on the micrographs represent time (s). For more examples, see Extended Data Fig. 2. (c) Top: Matrix representation of the organelle interactome. The absolute numbers of organelle contacts in a single cell at a single time point are displayed as a graphical half matrix. Each row in the matrix represents the number of organelle contacts with each target organelle (columns), and is colour coded from 0 to 600. Bottom: Organelle interactome over time. Each half matrix represents the organelle interactome in a single cell at a specific time point. (d) Network representation of the organelle interactome in all ten cells. All nodes (organelles) are connected and the length of the edges connecting two nodes represents the inverse of the number of contacts between those two organelles. Mito, mitochondria; perox, peroxisomes; lyso, lysosomes. Scale bars, (a) 10 μm, (b) 5 μm.
